Cotranslational microRNA mediated messenger RNA destabilization
Figures
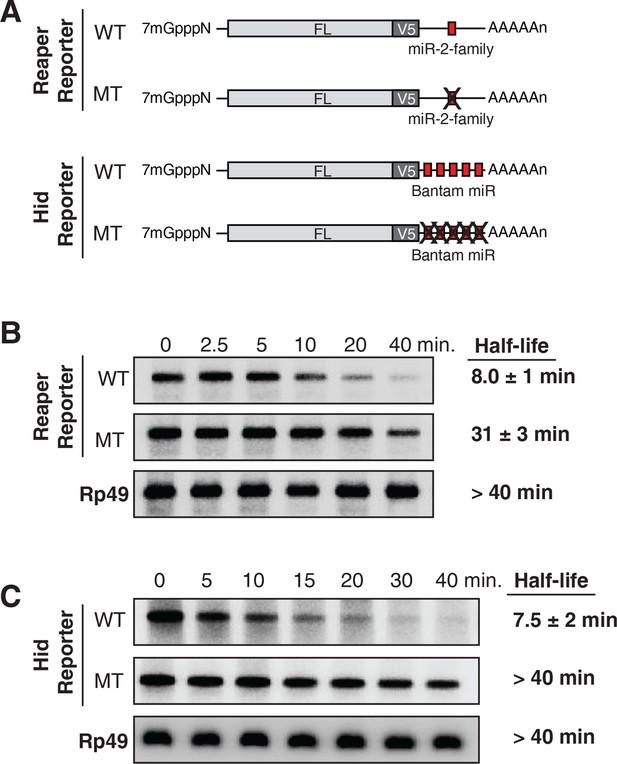
Destabilization of reaper and hid reporter mRNAs by endogenous miRNAs.
(A) Schematic of reporter mRNAs analyzed; WT denotes mRNAs with wild type MREs and MT denotes mRNAs where the MREs were inactivated by mutation. FL is the firefly luciferase coding region and V5 is the V5 epitope tag. (B) Stability of reaper reporter mRNAs containing wild type (WT) or mutant (MT) MREs. Cells were treated with actinomycin D for the indicated times; total RNA was prepared and analyzed by Northern blotting using a probe to the firefly luciferase open reading frame. Rp49 denotes the mRNA encoding ribosomal protein L32, a highly stable mRNA, which served as an internal loading control. (C) Stability of hid reporter mRNAs containing wild type (WT) or mutant (MT) MREs. All procedures were exactly as in (B).

Sequences of 3’ UTRs of reaper and hid with wild type and mutant MREs highlighted.
(A) Reaper 3’ UTR. The wild type MRE is shown in red while the mutant MRE is shown in blue. (B) Hid 3’ UTR. Wild type MREs for bantam are shown in red while mutant MREs are shown in blue.
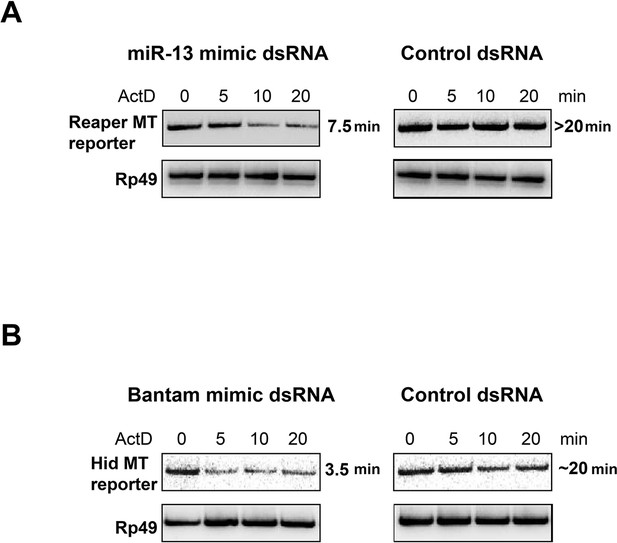
miRNA mimics complementary to mutant MREs destabilize reaper and hid reporter mRNAs.
For miRNA mimics, ds RNAs were ordered from Integrated DNA Technologies (IDT) according to their protocol for design of miRNAs. The mir2 mimic was synthesized to contain a seed sequence complementary to the altered nucleotides in the mutated reporter reaper reporter UTR. For the Bantam mimic, the seed was complementary to the new target sequence shown in supplemental Figure 1—figure supplement 1 present in all of the altered Bantam sites. The control dsRNA was a negative control recommended by IDT. For optimization of transfection conditions a double stranded siRNA of the same design was used to target the Firefly luciferase coding sequence. Titrations of dsRNA, plasmid and transfection reagent (Mirius Transit-X2) were performed to optimize expression and knockdown conditions as assayed by Luciferase readings in cells cotransfected with Firefly siRNA. For half life measurement, S2 cells were co-transfected with reporter and dsRNA in 10 cm plates at a starting density of 2X106/ml. 24 hr after transfection, cells were removed from plates, distributed on 6 well plates and 0.1 mM CuSO4 was added for an additional 16 hr at which point 5 µg/ml Actinomycin D was added, media was removed at the indicated times and cells were lysed with Trizol. Because of the low abundance of transfected reporter RNAs, the analysis of RNA half life was done by semi-quantitative reverse transcription PCR (Maroney et al., 2006). cDNA was synthesized from 0.5 µg total RNA using random primers and Superscript II. PCR reactions were first titrated for linearity using a series of cDNA dilutions and cycle numbers. Linear PCR reactions (25 cycles for Reaper and Hid reporters, 20 cycles for Rp49) were carried out in the presence of α32P dCTP and analyzed on non-denaturing 6% polyacrylamide gels.
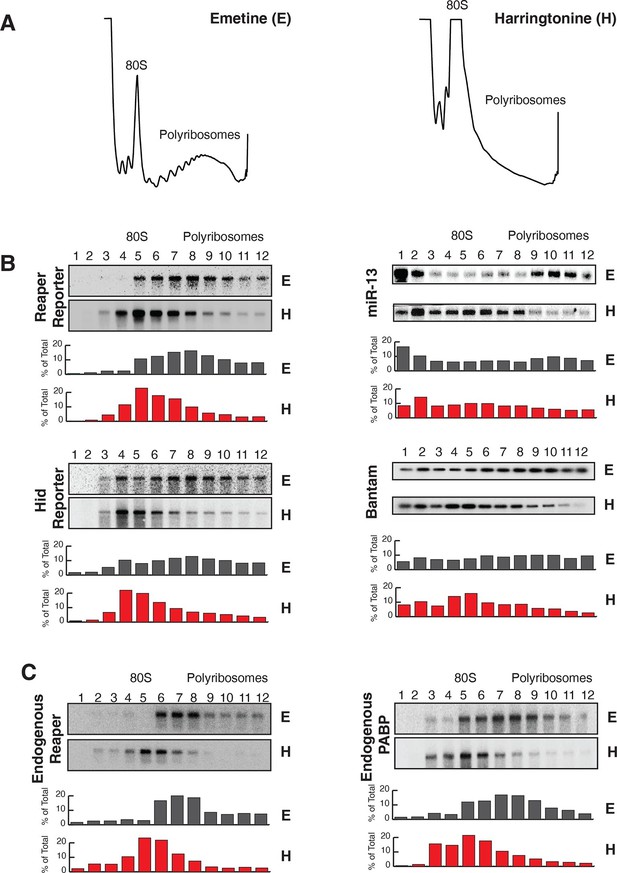
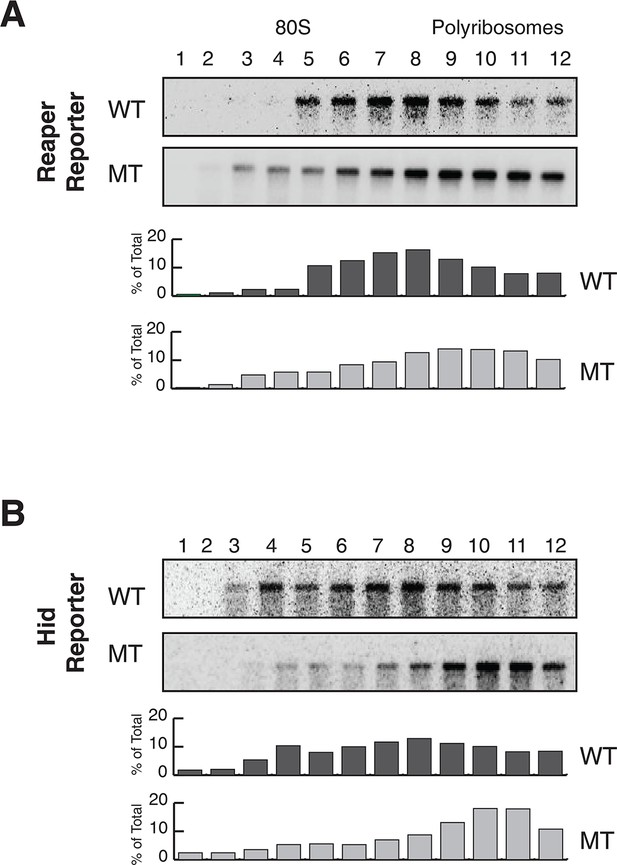
Reaper and hid reporter mRNAs containing wild type MREs are present on actively translating ribosomes.
(A) Representative UV absorbance (254 nm) traces of polysome gradients on which either cytoplasmic extracts from emetine (E) treated or harringtonine (H) treated cells were fractionated. (B) Northern blots and quantitation of signal from fractions of polysome gradients as in (A); Blots from cells expressing reaper and hid reporter mRNAs containing wild type MREs are on the left. Sedimentation profiles, as assessed by splinted ligation, and signal quantitation of endogenous mir-13b and bantam miRNAs are shown on the right. (C) Northern blot and quantitation of signal from fractions of polysome gradients in which extract of cells treated with emetine (E) or harringtonine (H) were sedimented. The probes were to either endogenous reaper or to polyA binding protein (PABP) mRNAs.
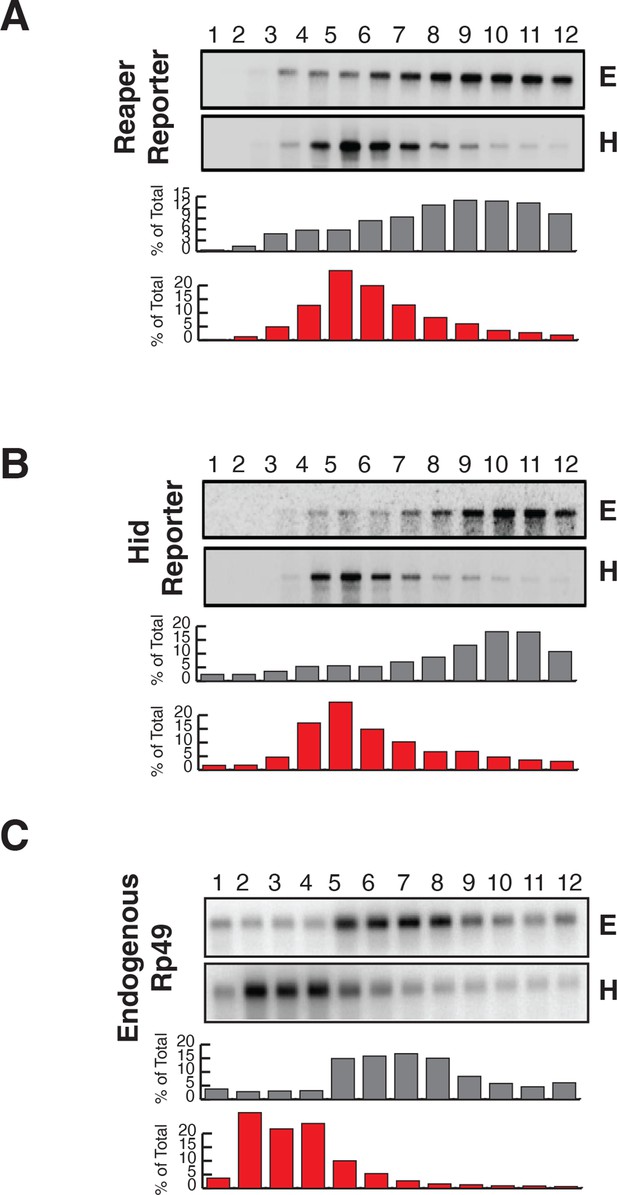
Reporter mRNAs containing mutant MREs are present on translating ribosomes.
(A) (B) and (C) are exactly as described for (A) (B) and (C) in Figure 2 except that reporters containing mutant MREs were analyzed.
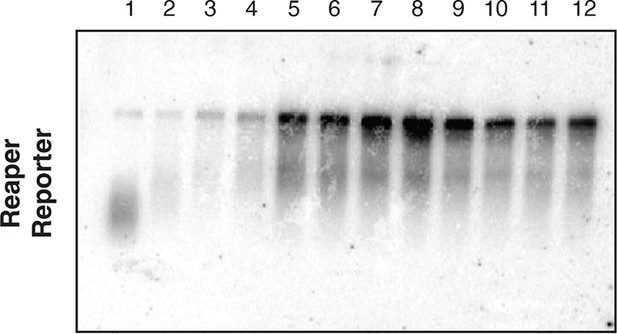
First indication of cotranslational decay of reaper reporter mRNA containing a wild type MRE.
Norther blot as shown in Figure 2 (A) was exposed for a longer time. Note fragments smaller than full length.
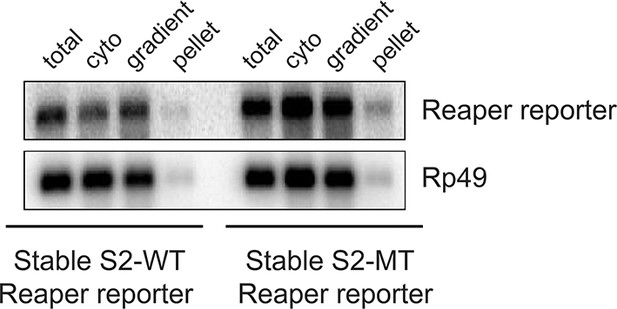
Most mRNAs are accounted for in polysome gradients.
We did an accounting experiment analogous to that shown in Maroney et al., 2006. Equal numbers of cell from wt or mutant reporter cell lines were processed as described in methods to prepare cytoplasmic (cyto) extract or to lyse with Trizol (total). RNA was then prepared from half of each sample. The remaining half of cytoplasmic extract was centrifuged through the same sucrose gradients used for polysome analysis. Fractions were pooled (gradient) and pellets (pellet) resuspended and RNA was prepared from both. After EtOH precipitation all samples were resuspended in the same volume. Equal aliquots of each were used in Northern blots and probed for both reporter and control (Rp49) mRNAs.
Reporter mRNAs containing wild type MREs are associated with lighter polysomes than reporter mRNAs containing mutant MREs.
(A) Direct comparison of sedimentation reporter mRNAs containing wild type WT or mutant MT MREs. These are the same blots and quantitations as those shown in a different context in Figure 2 and Figure 2—figure supplement 1. (B) Same as in (A) but hid reporter mRNAs were analyzed here.
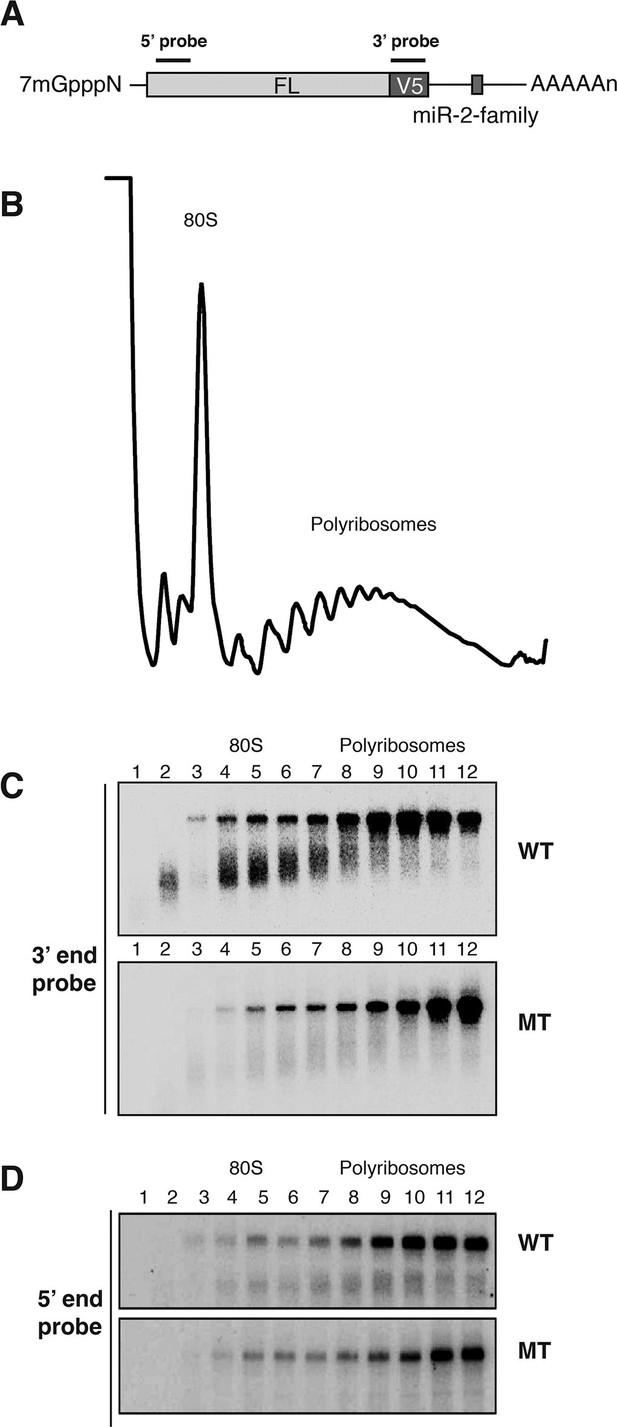
Cotranslational decay of reaper reporter mRNA.
(A) Schematic representation of probes used for detection of reaper reporter mRNA. (B) Representative UV absorbance trace (254 nm) of a polysome gradient fractionating cytoplasmic extract prepared from cells treated with actinomycin D for ten minutes. Actinomycin treatment for this time had no effect on overall protein synthesis as assessed by polysome profiling (Figure 3—figure supplement 1). (C) Northern blot analyses of fractions from polysome gradients in which cytoplasmic extracts from cells expressing reaper reporter mRNAs containing either a wild type (WT) or mutant (MT) MRE were sedimented. Northern blots were probed with a fragment that hybridized to the 3’ region of the firefly reporter open reading frame, see (A). (C) The same as in (D) except that the blots were probed with a fragment that hybridized to the 5’ region of the firefly luciferase open reading frame, see (A).
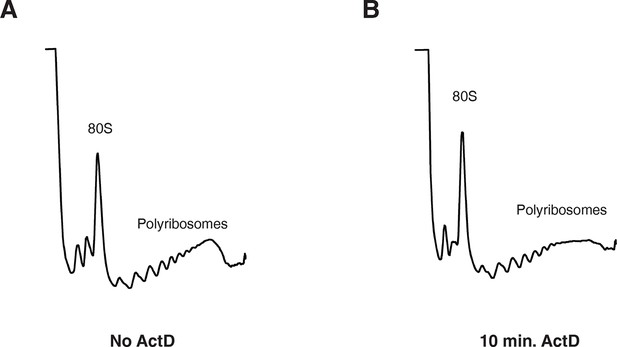
10 min actinomycin D treatment does not affect overall protein synthesis.
(A) UV absorbance (254 nm) trace of a polysome gradient using cytoplasmic extract from untreated cells. (B) same as (A) except that extract was prepared from cells treated with actinomycin D for 10 min.
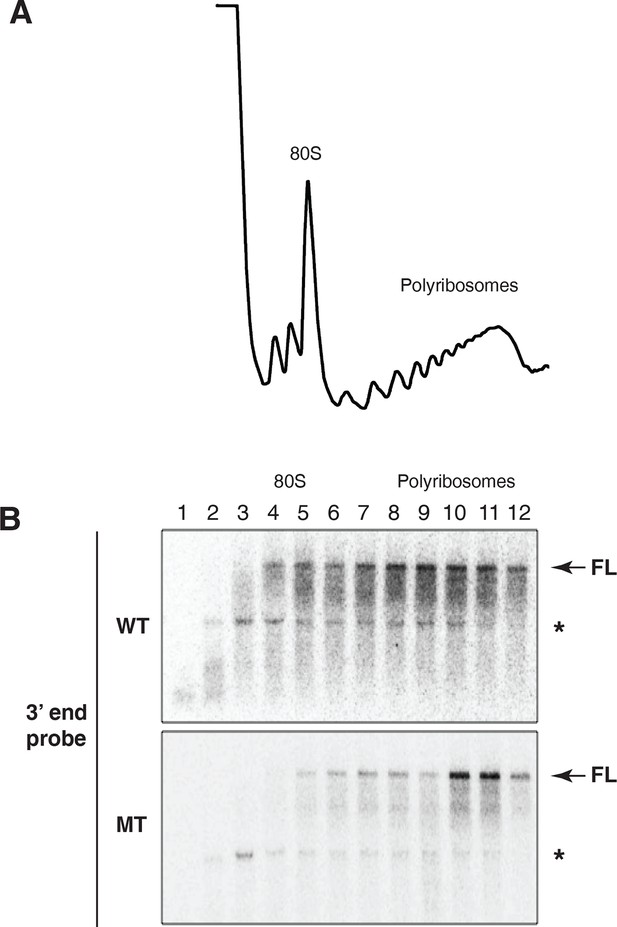
Evidence for cotranslational mRNA degradation of hid reporter mRNA containing wild type MREs.
(A) UV absorbance (254 nm) trace of polysome gradient fractionating cytoplasmic extract prepared from cells expressing the hid reporter construct containing wild type MREs. (B) Northern blots across polysome gradients of fractionated cytoplasmic extracts prepared from cells expressing hid reporter mRNAs containing wild type (WT) or mutant (MT) MREs. FL denotes full length mRNA. The asterisk denotes a band that comigrates with 18S RNA and represents some trapping of hid reporter mRNA fragments. Probe was 3’ probe in Figure 3A.
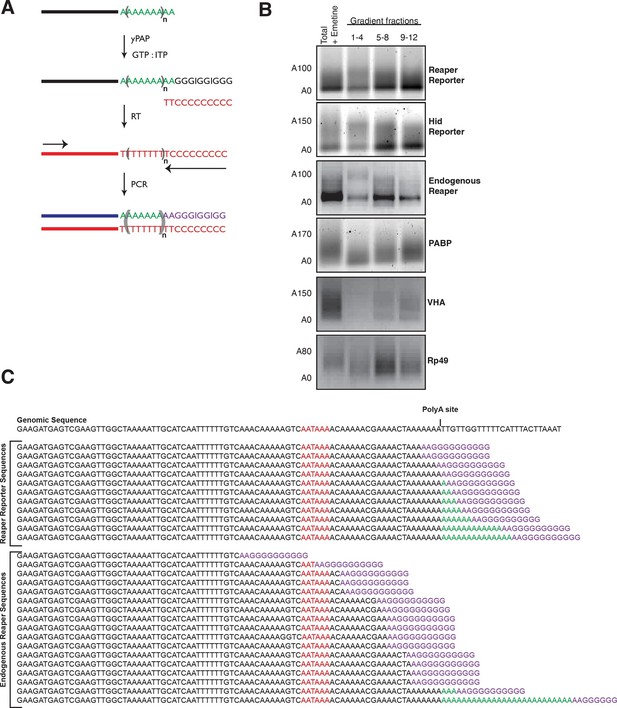
Cotranslational miRNA-mediated decay involves mRNA deadenylation.
(A) Schematic depiction of the polyA tail length assay to measure the length of a polyA tail. (B) PolyA tail lengths for the indicated mRNAs. Tail lengths were determined for each mRNA in total RNA and pooled polysome gradient fractions as indicated. This assay does not have the sensitivity to detect the endogenous hid 3’ end. (C) The deadenylated species of reaper reporter and endogenous reaper were cloned and sequenced. AATAAA: polyA signal. Genomic DNA sequences are shown in black while the reverse primer is shown in purple.
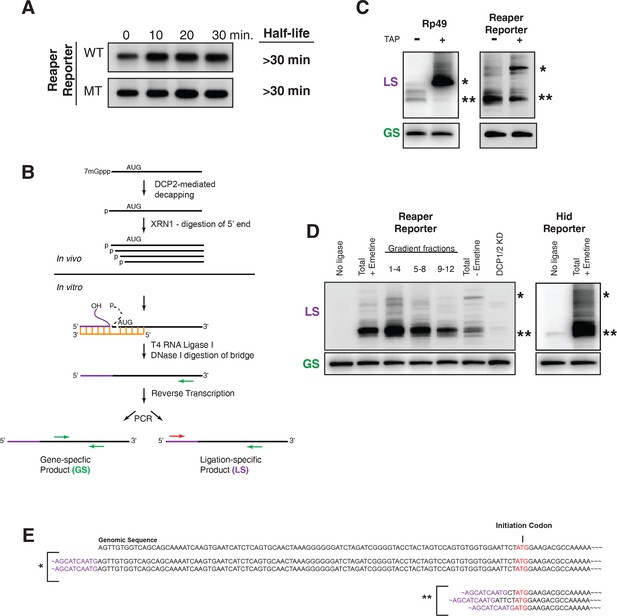
Cotranslational miRNA-mediated decay involves mRNA decapping.
(A) Stabilization of reaper reporter mRNAs upon knock down of Drosophila Dcp1 and Dcp2. Northern blot analysis of total RNA prepared at the indicated time after addition of actinomycin D and probed with a fragment that hybridized to the 3’ region of the firefly luciferase open reading frame. (B) Schematic depiction of the ligation strategy to identify decapped 5’ phosphorylated mRNA fragments. (C) Ligation assay for the indicated mRNAs either untreated or treated with tobacco acid pyrophosphatase (TAP). Ligation specific (LS) (decapped RNA) or gene specific (GS) PCR fragments; see (B), Heterogeneity results from digestion by XRN1 following decapping. The single asterisk denotes the position of the product when ligation is at the 5’ terminus of the mRNA. The double asterisk denotes ligation products near the AUG initiation codon. (D) Ligations assays on RNA samples from cytoplasmic extract and gradient fractions, or total RNA with or without knock down of decapping enzymes. Labelling is as in (C). (E) Sequencing analysis of the products when ligation is to the 5’ terminus of the mRNA and near the ATG initiation codon. Ligation-specific forward primer is shown in purple.
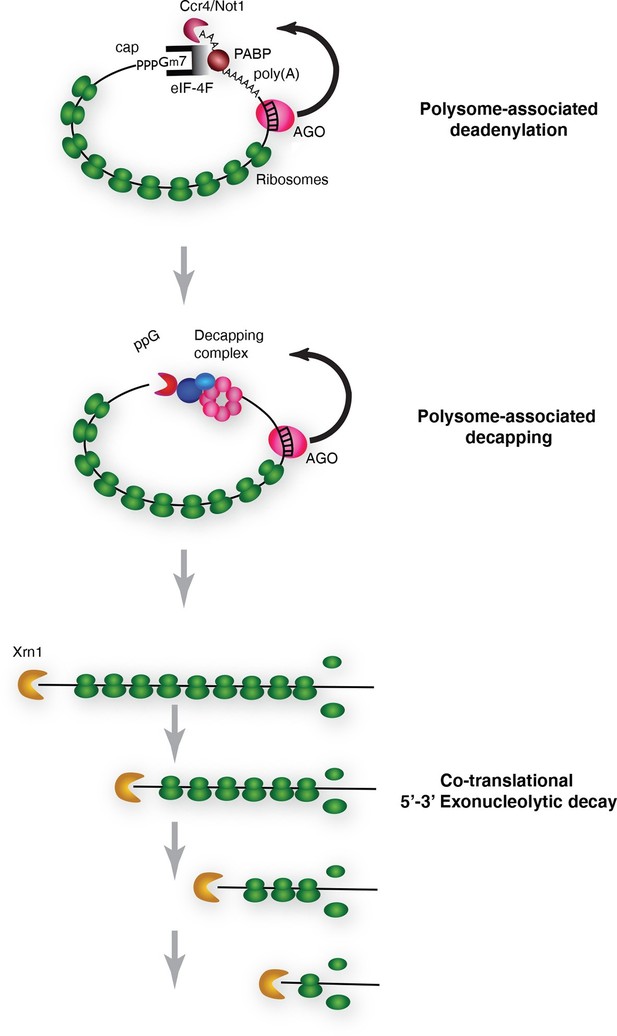
Pathway of miRNA-mediated cotranslational mRNA decay.
Schematic summary of the model which emerged from the data; for details see the text.
Additional files
-
Supplementary file 1
Oligonucleotides used in this study.
- https://doi.org/10.7554/eLife.12880.017





