Synthetic protein interactions reveal a functional map of the cell
Figures
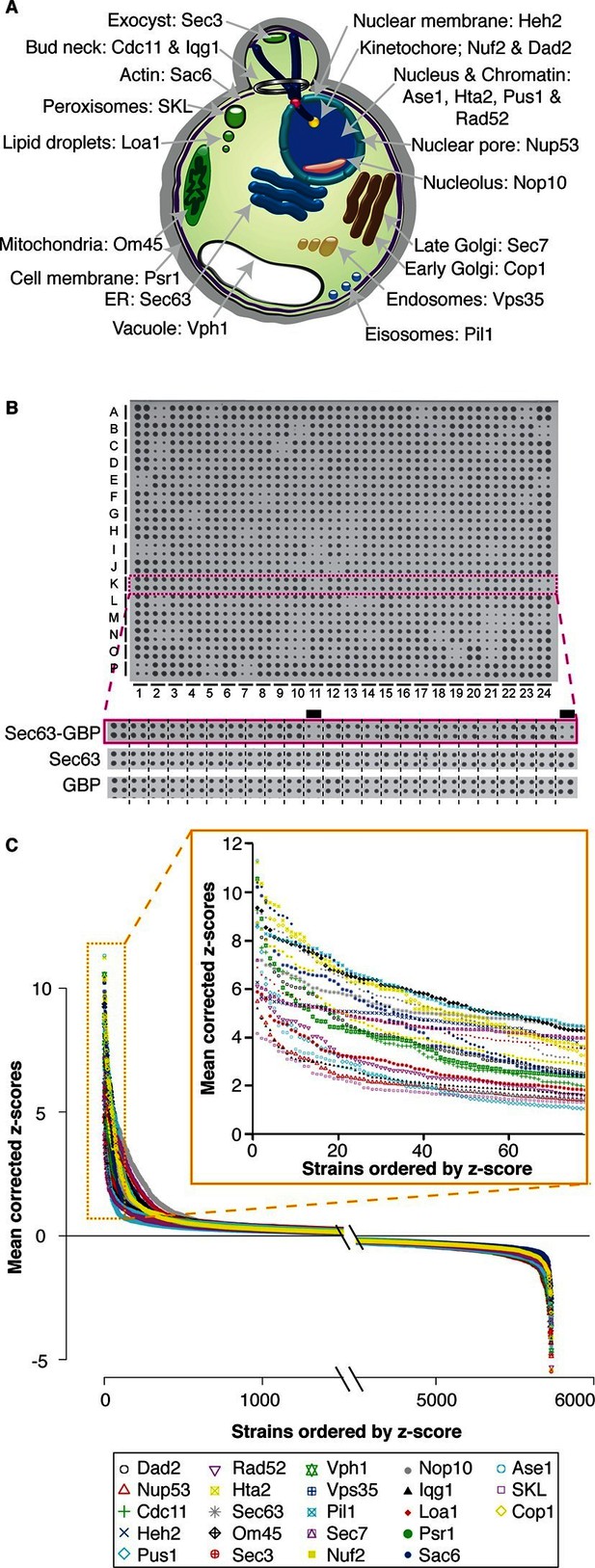
Quantitative analysis of the effects of binding proteins throughout the cell.
(A) A schematic diagram of S. cerevisiae indicating the cellular compartments and target proteins within the cell that were associated with each member of the proteome. (B) A 1536 colony plate from the Sec63 screen. The inset below shows the highlighted row from the Sec63-GBP plate, the Sec63 control plate and the GBP-only control plates respectively. Growth defects are indicated with a black line. (C) The z-scores of all 5734 proteins in each of the 23 screens. For each screen, the strains are ranked according to order of z-scores, positive z-scores indicate a growth defect relative to controls. The inset highlights the strains with the largest growth defects in each screen.
-
Figure 1—source data 1
Z-scores of growth defects caused by protein associations.
The z-scores for each binary protein interaction are listed for each control. The mean smoothed z-scores, plotted in panel C, are listed separately.
- https://doi.org/10.7554/eLife.13053.004
-
Figure 1—source data 2
Plasmids used in this study.
A list of the plasmids used to generate the SPI data are listed.
- https://doi.org/10.7554/eLife.13053.005
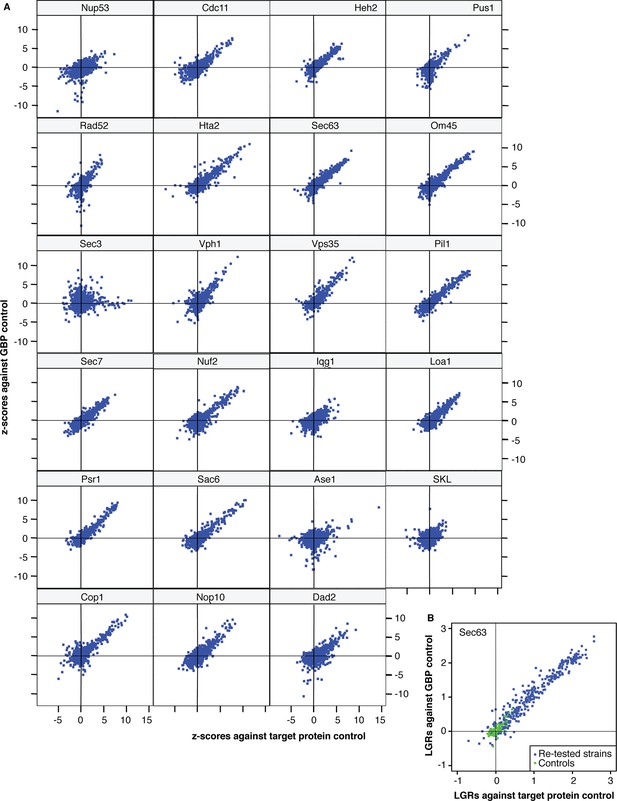
The correlation between the two controls.
(A) x-axis: z-scores quantifying the difference in colony sizes between strains expressing the target protein fused with GBP and strains expressing the target protein alone, to control for the effects of ectopic expression. y-axis: z-scores quantifying the difference in colony sizes between strains expressing the target protein fused with GBP and strains expressing the GBP alone, to control for the effects of binding a protein to the GFP tag. A positive z-score indicates a growth defect of the GBP-tagged target protein relative to its control. (B) The relative growth (Log Growth Ratios, LGRs) from the high-density re-testing of Sec63-GBP SPIs relative to the two controls (the target protein alone – x-axis and the GBP alone – y-axis). The relative growth of the negative controls (no GFP tagged protein) are shown in green, whereas the retested Sec63 SPIs are shown in blue.
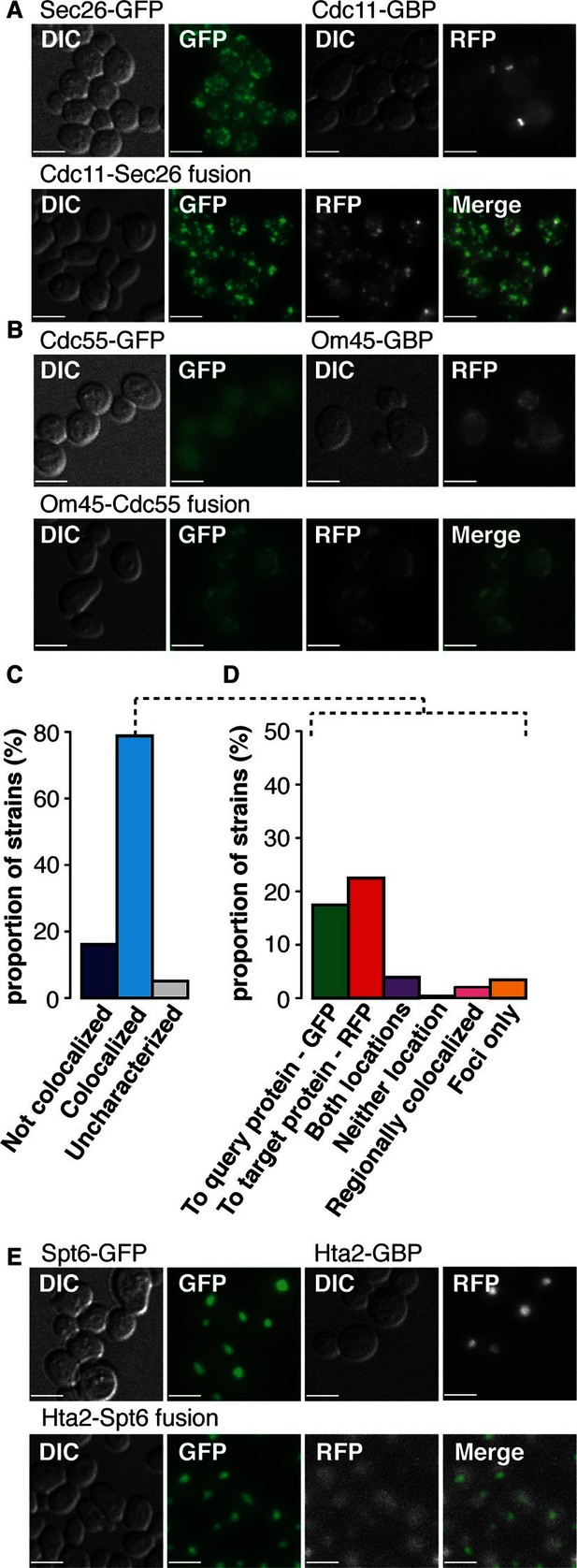
Colocalization of target GBP protein and query GFP proteins.
(A) Cdc11-GBP relocalizes to the Golgi when bound to Sec26-GFP. (B) Cdc55-GFP relocalizes to the mitochondria when bound to Om45-GBP. (C) Bar chart of the proportion of colocalization (n=552), note that the colocalized category includes 210 combinations where the target and query proteins are within the same compartment and so protein-protein association will not be apparent from this microscopy analysis. (D) Bar chart of the direction of movement of GFP and GBP (n=225). ‘To query protein - GFP’ indicates relocation of the majority of the GBP target protein to GFP (see A); ‘To target protein - RFP’ denotes relocation of the majority of the GFP query proteins to the GBP-RFP target (see B). ‘Both locations’ indicates that GBP and GFP proteins are in both their normal location and those of the other protein (e.g. Figure 2—figure supplement 1B). ‘Neither location’ denotes both GFP and GBP proteins are colocalized, but not to either of their normal locations (e.g. Figure 2—figure supplement 1C), whereas ‘Regionally colocalized’ indicates one protein is in the same region of the cell as the second protein, but not completely colocalized (see E). ‘Foci only’ designates that the proteins relocalized to discrete foci (see Figure 2—figure supplement 1E). Two categories are omitted from this analysis, first those cells which were uncharacterized, typically because the cells were dead. Second, cells in which the target and query protein reside in the same cellular location, such that microscopy is not informative on whether or not they are associated, this latter category make up ~40% of our combinations. (E) Hta2-GBP is displaced from the nucleus when bound to Spt6. The scale bars are 5 µm.
-
Figure 2—source data 1
Localization summary.
These data provide a detailed list of the localization type for each GBP-GFP protein pair.
- https://doi.org/10.7554/eLife.13053.008
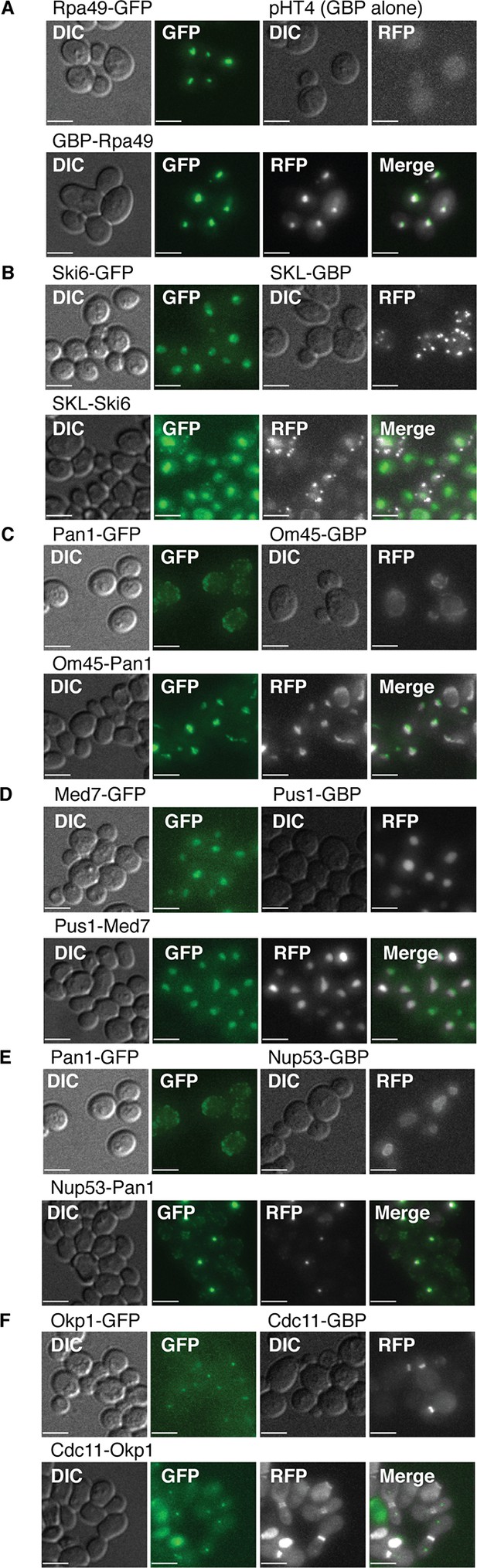
Fluorescent images of colocalization between GBP and GFP.
(A) GBP cololcalizes with Rpa49-GFP. (B) Both GFP and RFP can be detected in the nucleus and peroxisomes when Ski6-GFP and SKL-GBP bind. (C) Pan1-GFP and Om45-GBP colocalize in neither of their expected locations. (D) Med7-GFP and Pus1-GBP both localize to the nucleus, so displacement due to colocalization is impossible to detect. (E) Pan1-GFP and Nup53-GBP colocalize to form foci in the cell. (F) Okp1-GFP and Cdc11-GBP do not colocalize. The GFP kinetochore signal and RFP bud neck signal are still clearly visible in the combined strain. Scale bars are 5 µm.
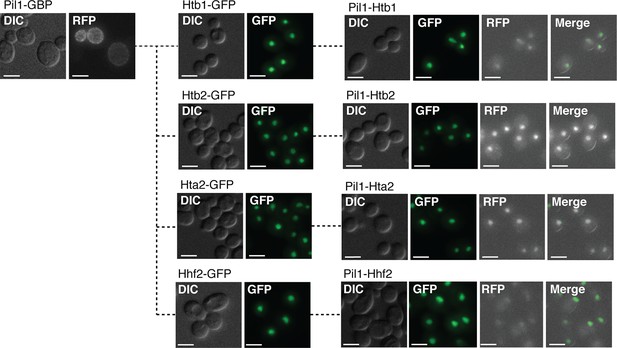
Colocalization of Pil1-GBP with histone subunits.
Localization of PIL1-GBP-RFP is shown, along with Htb1-GFP, Htb2-GFP, Hta2-GFP, and Hhf2-GFP. DIC, GFP, RFP and merged images then show the colocalization of each histone subunit (GFP) with Pil1-GBP-RFP. Scale bars are 5 µm.
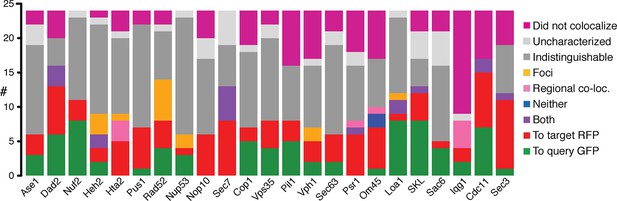
Direction of colocalization.
(A) The proportion of the 24 query proteins that colocalized in the direction indicated. Categories used to characterize the direction of colocalization are described in Figure 2. The ‘Uncharacterized’ category includes strains where there were no cells to image, which is often the case if the interaction perturbs growth.
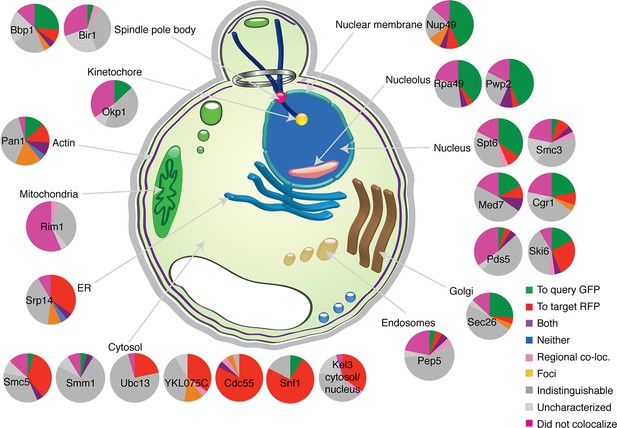
Direction of colocalization.
A pie chart for each of the 24 randomly selected query proteins shows the proportion of the 23 target proteins observed to have colocalized in the direction indicated. Categories used to characterize the direction of colocalization are described in Figure 2.
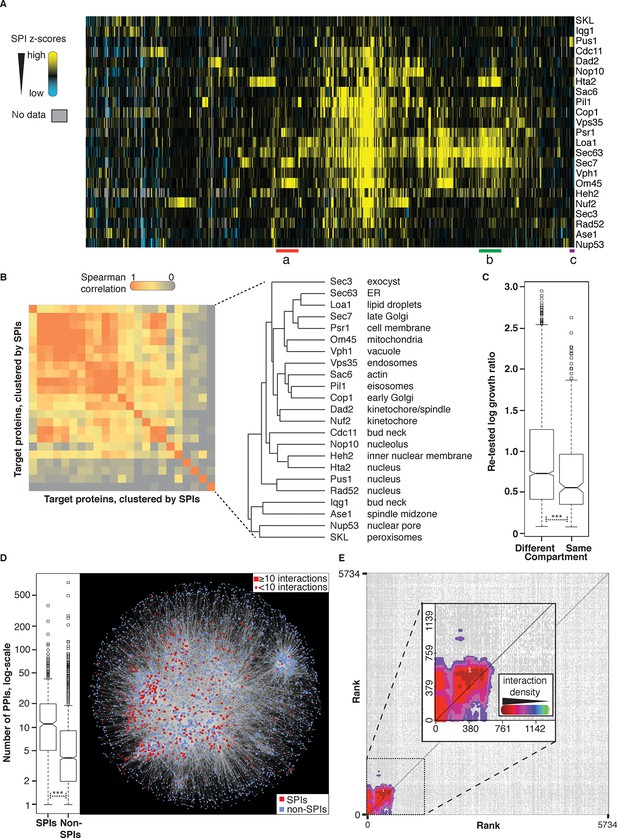
Comparisons of synthetic physical interaction screens.
(A) Cluster analysis of the SPI data. The 23 screens are arranged horizontally and the 727 GFP strains clustered vertically. High z-scores (positive; >2) in yellow and low (negative; < -1) scores in blue. Three distinct clusters are highlighted (a, b, and c) and described in Figure 4—figure supplement 6. (B) Spearman’s Rank Correlation Coefficients for the different SPI screens shows similar compartments give similar SPIs, for example, Sec63 and Loa1 cluster together as do two kinetochore proteins Nuf2 and Dad2. (C) The notched box-and-whisker plot indicates the distributions of the retest log growth ratios and indicates that SPIs produced by a query protein and a target protein from different compartments produce stronger growth defects than those from the same compartments (***indicates a p-value = 1.8x10-5, Wilcoxon's rank-sum). The plot shows the median value (bar) and quartiles (box), the whiskers show the minimum of the range or 1.5 interquartile ranges, outlying data points are indicated as circles and the notches indicate the 95% confidence intervals of the medians. (D) The GFP proteins with SPIs have, on average, more protein-protein interactions than non-SPI query proteins, the notched box-and-whisker plot is in the same format as panel B (***indicates a p-value <.2x10-16, Wilcoxon’s rank-sum). The 727 SPI query proteins (red) are superimposed upon the yeast interactome with proteins with ≥10 interactions shown as larger squares. (E) The CLIK interaction density plot for Sec63 is shown (see Figure 4—figure supplement 5 for the other CLIK plots). The ~500 Sec63 associations that show the strongest growth restriction have a high interaction density (inset).
-
Figure 4—source data 1
High-density retesting of the SPIs.
All the high-density SPI data to retest the strongest interactions are listed together with a list of those GFP strains that produce a reproducible SPI with each of the 23 GBP proteins. We also include a list of the growth data for a subset of GFP proteins whose expression and location is well characterized.
- https://doi.org/10.7554/eLife.13053.014
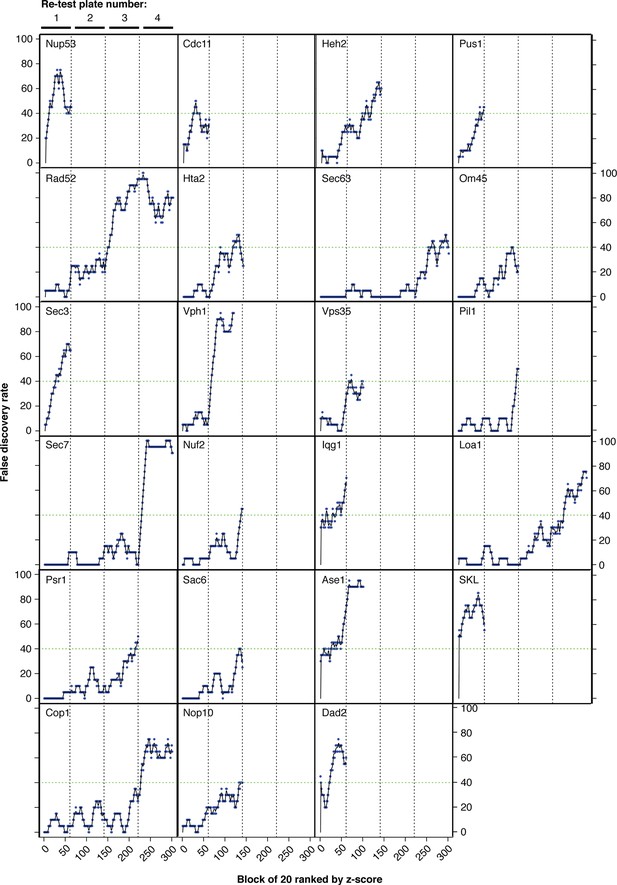
False discovery rates (FDR).
Strains were ordered from highest to lowest z-score and the strongest 80 growth defects were retested with 16 replicates (80 strains with 16 controls per plate). The FDR was calculated for each batch of 20 strains working down the list of z-scores, shown here in blue. Black lines indicate three-point moving averages. Vertical dashed lines group points from each retest plate together. No more retests were performed once a screen had reached 40% FDR, indicated by a horizontal green dashed line.
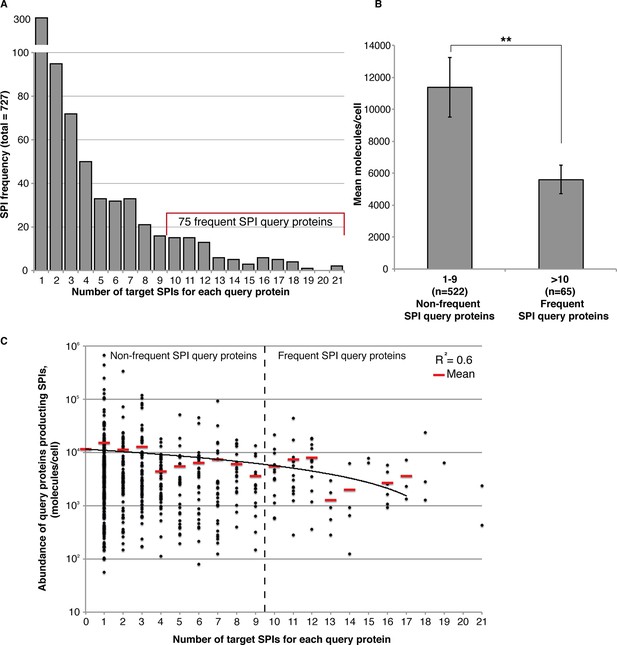
Frequent SPIs are with lower abundance query proteins.
(A) The distribution of SPIs is shown as a bar chart illustrating that most SPI query proteins give a SPI with only one or a few target proteins. Nevertheless, 75 query proteins have SPIs with at least 10 target proteins, the ‘Frequent SPIs’. (B) Frequent SPI query proteins have a lower mean abundance than non-frequent SPI query proteins (t-test, p>0.006). Error bars indicate standard error of the mean. (C) Frequency data shown in (A) plotted in terms of protein abundance. The mean abundance values in each category are indicated as red lines a linear trendline is shown in black.
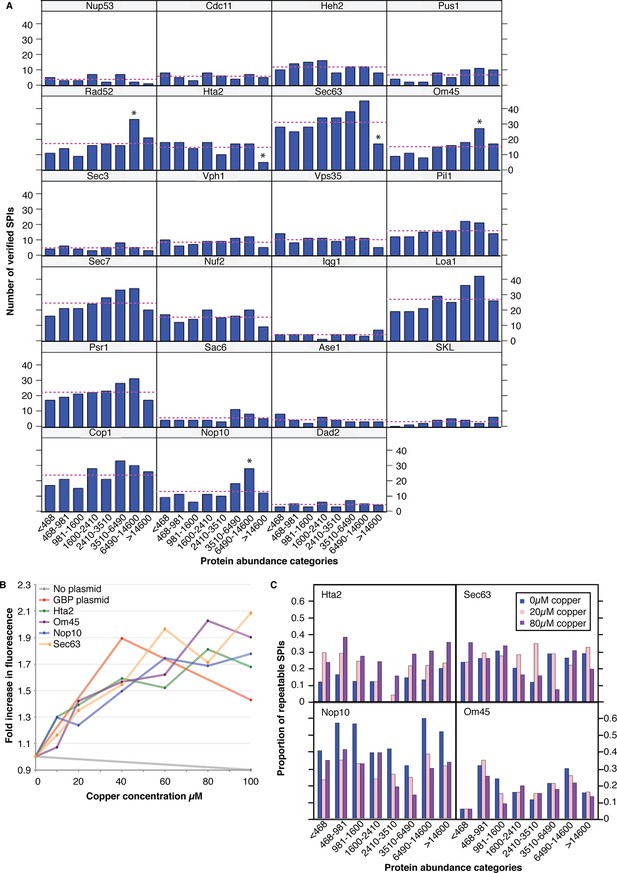
The effect of protein abundance on the number of verified SPIs.
(A) The number of verified SPIs in each abundance category is shown for each screen, where abundance categories are bins containing an equal number (421) of proteins. The dashed line indicates the number of SPIs expected per category if distribution was entirely unbiased. (B) GBP-tagged protein levels were assessed via RFP fluorescence. Addition of copper to the medium increased the total protein concentration by as much as twofold. (C) 400 GFP strains were chosen to provide representatives in each protein abundance category, with a bias toward those in the highest abundance category. These GFP strains were retested with the four GBP-tagged proteins at different copper concentrations (0, 20, and 80 µM) and the proportion of SPIs within each category are indicated. Addition of copper did not increase the proportion of SPIs specifically with the high-abundance protein categories. It is of note that increasing the amount of Hta2-GBP produced more SPIs in all abundance categories, whereas increasing the amount of Nop10-GBP reduced the number of SPIs in all categories. Hence, although the amount of GBP protein can affect the SPIs, it does not correlate with GFP protein levels.
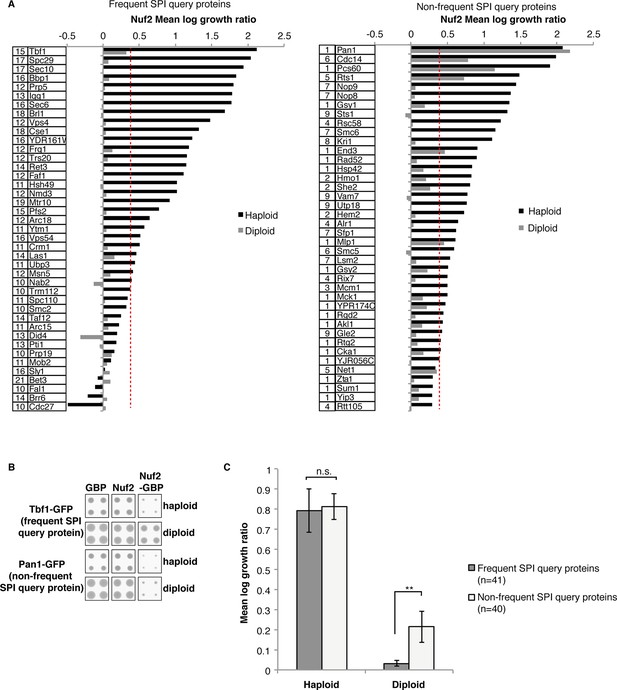
Frequent SPIs are rarely dominant.
(A) From the Nuf2 SPI screen 41 frequent SPI query proteins (left panel) and 40 non-frequent SPI query proteins (right panel) were retested both as haploids and diploids. The numbers in the box to the left of the ORF name indicate the number of screens, out of 23, that the GFP-query protein was detected as a SPI. All the frequent SPI query proteins were not dominantly restricted for growth when associated Nuf2, compared with 15% (6/40) of the non-frequent SPI query proteins. (B) An example of the raw data with one frequent SPI (Nuf2-Tbf1) and one non-frequent SPI (Nuf2-Pan1) illustrate the suppression of the former SPI in diploid cells. (C) The mean log growth ratio of the combined 41 frequent and 40 non-frequent SPI query proteins from the Nuf2 SPI screen shows that there is no difference between frequent and non-frequent haploid SPI query proteins, in contrast the frequent diploid SPI query proteins are suppressed compared to non-frequent diploid SPI query proteins. Error bars indicate standard error of the mean, and **indicates a t-test p-value of 0.006; n.s. indicates not statistically significant.
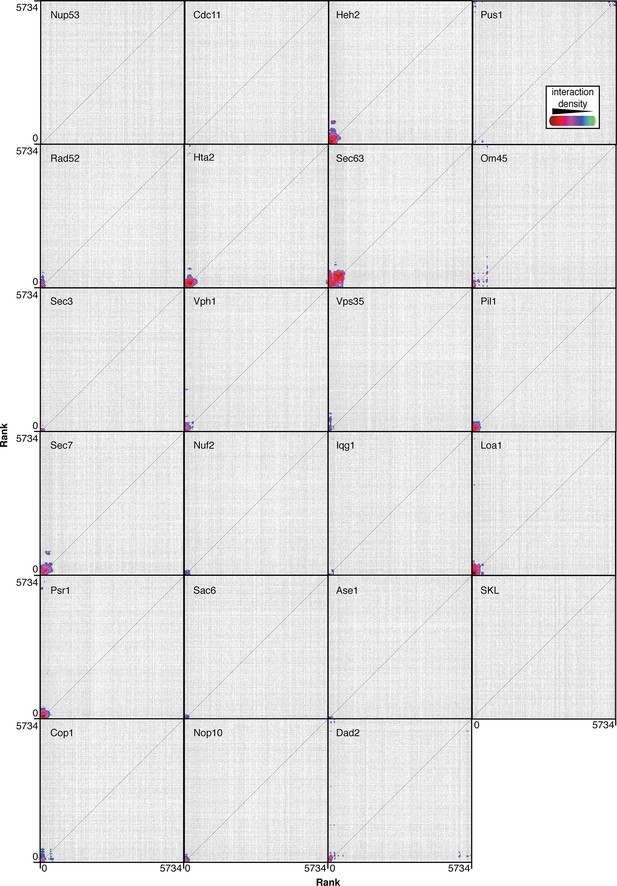
CLIK (Cutoff Linked to Interaction Knowledge) outputs for each of the 23 screens.
Strains are ranked according to z-score and plotted in this order along the x- and y- axes (at position 0 is the strain with the strongest growth defect). Points are plotted where a genetic or physical interaction exists between two strains, and colors indicate high density of interactions.
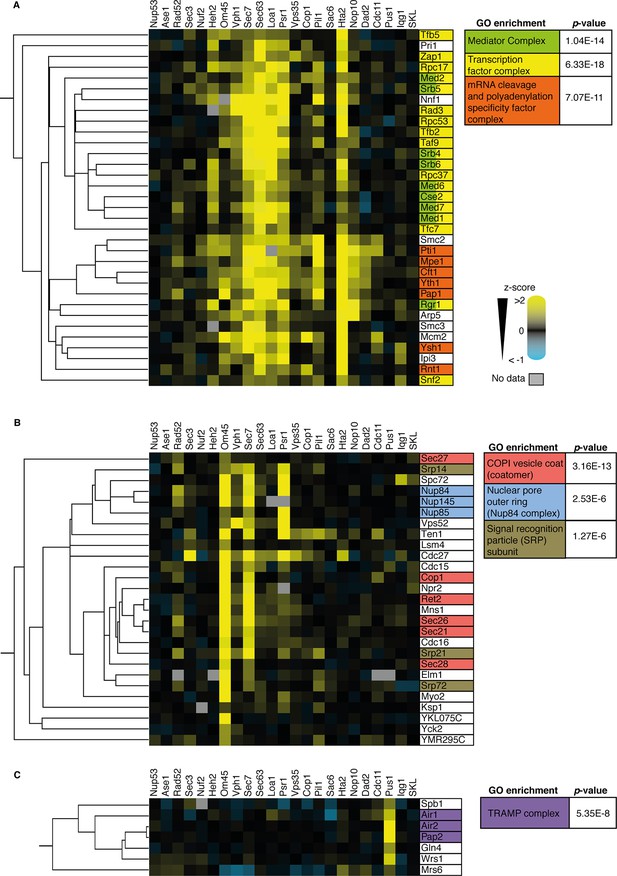
Analysis of three sub-clusters from the 727 SPI heat-map from Figure 3A.
(A) Many components of the mediator complex, transcription factor complex, and mRNA cleavage and polyadenylation specificity factor complex cluster together the SPI data (cluster a in Figure 4A). (B) Almost all components of the COP1 vesicle coat (or coatomer), nuclear pore outer ring (specifically the NUP84 complex), and signal recognition particle (SRP) subunits cluster together (cluster b in Figure 4A). (C) Members of the TRAMP complex cluster together and are specifically SPIs with the target protein Pus1 (cluster c in Figure 4A). p-Values for gene ontology (GO) term enrichments are indicated.
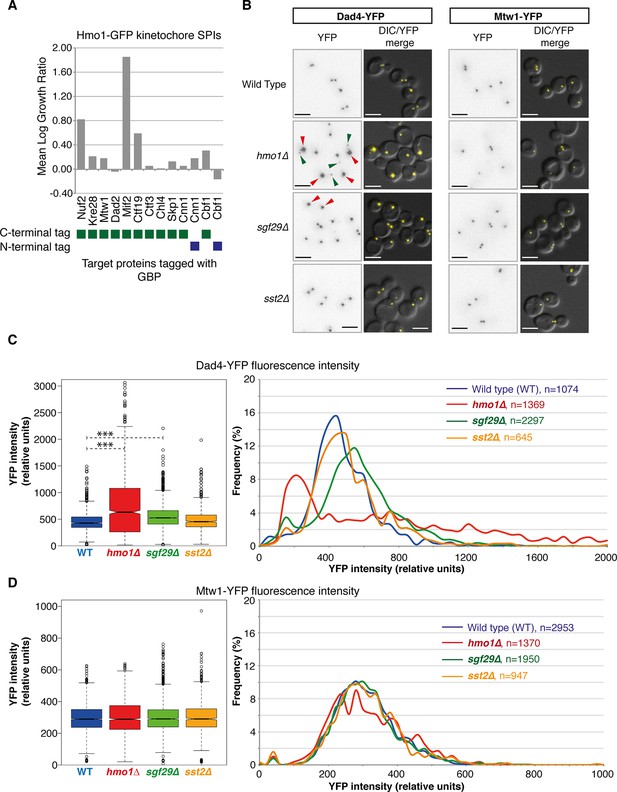
Nuf2 SPIs affect kinetochores.
(A) The Hmo1-GFP query protein encoding strain was transformed separately with 13 plasmids encoding different kinetochore proteins target proteins tagged with GBP (4 replicates each). The growth relative to controls (GBP alone and target protein alone) was assessed as in Figure 1. (B) Deletion of HMO1, SGF29, and SST2 were separately introduced into strains encoding Dad4-YFP and Mtw1-YFP at their endogenous loci. Fluorescence imaging of these strains reveals that hmo1∆ mutants have large-bright Dad4-YFP kinetochore foci (red arrows) and some weak foci (green arrows). sgf29∆ mutants contain bright Dad4-YFP foci (red arrows). In all cases, there are no effects upon Mtw1-YFP foci (right panels). Scale bars in all images are 5 µm. (C) Quantitation of the Dad4-YFP kinetochore foci fluorescence levels from these cells indicates that the levels of Dad4-YFP at kinetochores are affected by deletion of either HMO1 or SGF29. The left notched box and whiskers plot indicates the median (background subtracted) fluorescence values of kinetochore foci in relative units. The plot shows the median value (bar) and quartiles (box), the whiskers show the minimum of the range or 1.5 interquartile ranges, outlying data points are indicated as circles (note that several outlying data points are not shown as they are beyond the scale of the plot). The notches indicate the 95% confidence intervals of the medians (***indicates p-values <10–10 from a Wilcoxen’s rank-sum test). It should be noted that the distribution of kinetochore intensities do not conform to a normal distribution, particularly for the hmo1∆ mutant. The right panels show the distribution of fluorescent intensities of kinetochore foci of the same data plotted to the left (note that several outlying data points are beyond the scale of the plot). These data indicate the abundance of both the low and high intensity Dad4 foci of the hmo1∆ mutant (green and red arrowheads in Figure 5A, respectively) (D) Mtw1 kinetochore foci fluorescence levels are plotted as in panel C, we could not detect a difference from wild type cells in all three mutants.
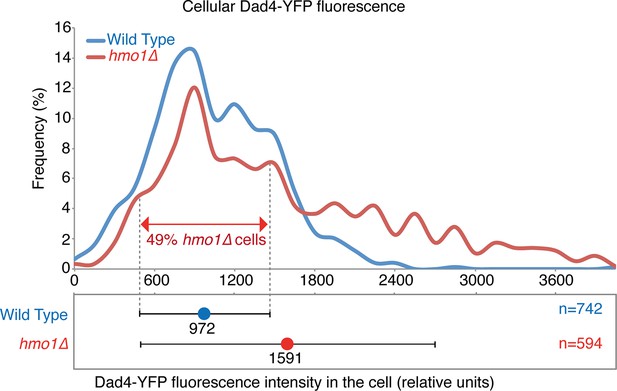
Cellular levels of Dad4-YFP in wild-type and hmo1∆ cells.
The histogram indicates the levels of Dad4-YFP fluorescence (relative units, r.u.) in both wild-type (blue) and hmo1∆ (red) cells. The plots below show the mean cellular fluorescence (wild type=972 r.u. and hmo1∆=1591 r.u. and the error bars indicate the standard deviation of the mean). The dashed lines indicate + and – one standard deviation of the mean of wild-type cells.
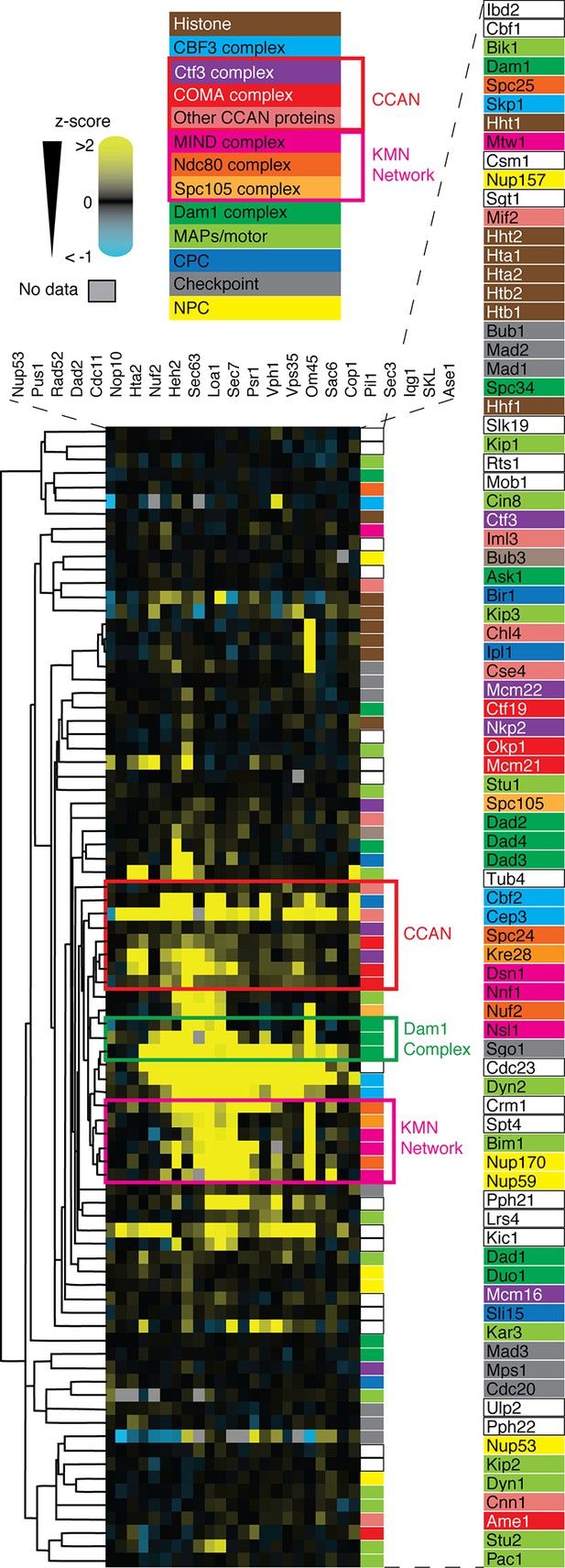
Cluster analysis of kinetochore and associated proteins using the SPI data are plotted as a heat-map.
High z-scores (positive; >2) are shown in yellow and low (negative; < -1) scores in blue (as in Figure 4A). The different protein complexes within the kinetochore are color-coded as indicated in the legend. Based on the SPI data alone, key complexes within the kinetochore cluster together as indicated by the colored boxed regions of the plot.
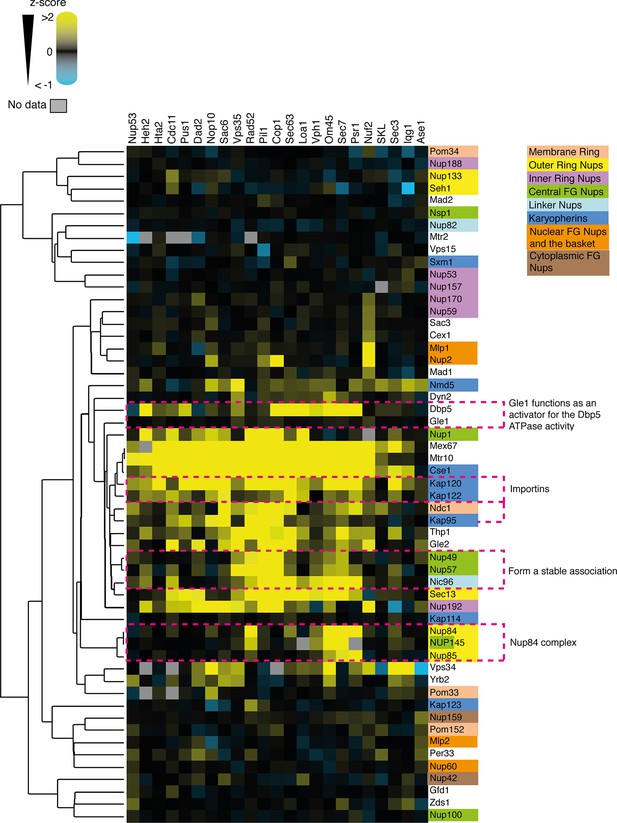
Clustering analysis of nuclear pore complex (NPC).
The SPI data are sufficient to cluster NPC subunits and karyopherins into some of the key functional complexes, such as the Nup84 complex of the outer ring of the nuclear pore. The data also cluster Gle1 with Dbp5. Gle1 normally regulates the activity of Dbp5. z-scores of NPC and NPC-associated GFP strains in all of the SPI screens are plotted as a heat-map. High z-scores (positive; >2) in yellow and low (negative; < -1) z-scores in blue. Both the screens and the GFP-tagged genes are clustered as in Figure 4A.
Tables
Yeast strains used in this study.
| Strain name | Genetic background | Relevant genotype | Reference |
|---|---|---|---|
| W8164-2B | W303 | MATα CEN1-16::Gal-Kl-URA3 | (Zou and Rothstein, 1997) |
| GFP strains | BY4741 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 XXX-GFP::HIS3 | (Huh et al., 2003) |
| PT147-7C | W303 | MATa TRP1 lys2∆ DAD4-YFP::NAT SPC42-RFP:: | This study |
| PT12-13D | W303 | MATa TRP1 MTW1-YFP hmo1∆::KAN | This study |
| T403 | W303 | MATa TRP1 lys2∆ DAD4-YFP::NAT SPC42-RFP::HYG hmo1∆::KAN | This study |
| T404 | W303 | MATa TRP1 lys2∆ DAD4-YFP::NAT SPC42-RFP::HYG sgf29∆::KAN | This study |
| T402 | W303 | MATa TRP1 lys2∆ DAD4-YFP::NAT SPC42-RFP::HYG sst2∆::KAN | This study |
| T406 | W303 | MATa TRP1 MTW1-YFP hmo1∆::KAN | This study |
| T407 | W303 | MATa TRP1 MTW1-YFP sgf29∆::KAN | This study |
| T405 | W303 | MATa TRP1 MTW1-YFP sst2∆::KAN | This study |





