Structural characterization of ribosome recruitment and translocation by type IV IRES
Figures
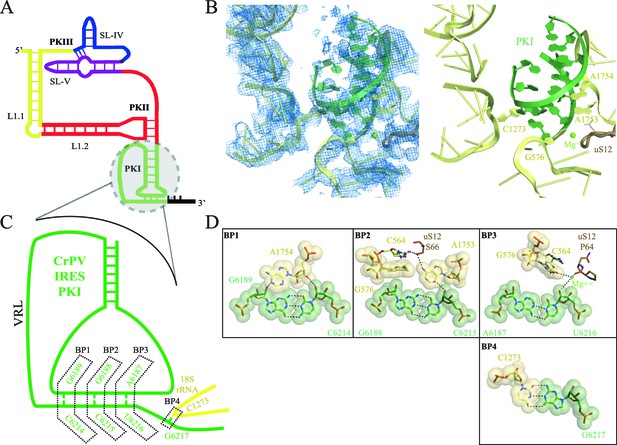
Interaction of PKI of CrPV-IRES in the 40S mimics decoding.
(A) Schematic representation of the type-IV IRES, colored according to different structural motifs. (B) Overall view of the interaction of the CrPV-IRES PKI (colored as in (A) in the context of the small ribosomal subunit (40S, yellow). On the left, experimental density is shown in the same orientation as on the right, where the final refined model is depicted. (C) Schematic view of PKI with main components labelled. (D) Space-filling models for the base pairs established between the anticodon and codon mimics of PKI (BP1-3) as well as the adjacent base pair (BP4).
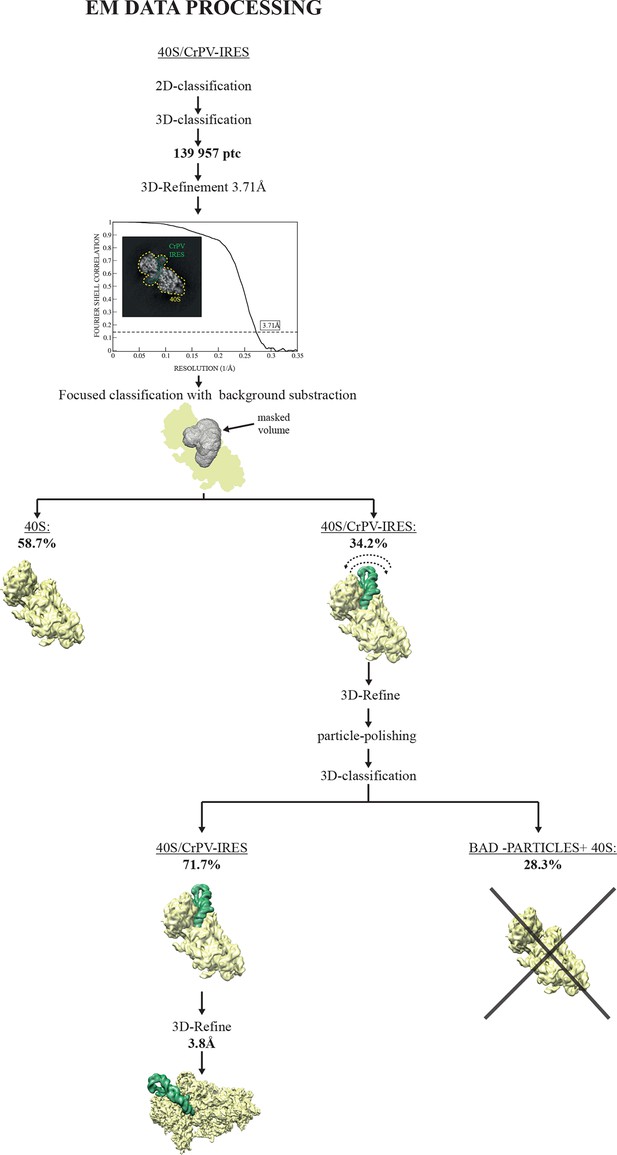
EM image processing workflow for the CrPV-IRES/40S data.
https://doi.org/10.7554/eLife.13567.003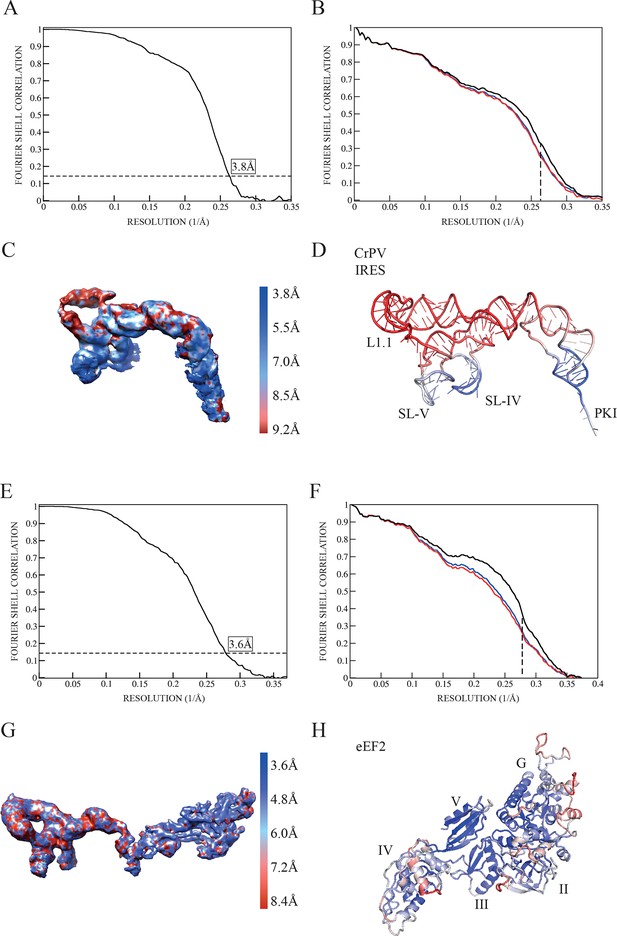
Quality of maps and models.
(A) Fourier shell correlation (FSC) of two independent half-maps for the complex CrPV-IRES/40S from the final refined class of particles. (B) FSC calculated for the refined model and the final map (black line). For cross-validation, the FSC calculated against a model refined with a half map (red line) is compared with the FSC calculated with the half map not included in the refinement (blue line). The overlapping of both lines provides evidence for the absence of overfitting in the refined model. (C) Unsharpened map colored according to the computed local resolution for the final map for the CrPV-IRES in the binary CrPV-IRES/40S complex. (D) Refined model of CrPV-IRES colored according to the atomic B-factor values (blue low, red high). (E) Fourier shell correlation (FSC) of two independent half-maps for the complex 80S/CrPV-IRES/eEF2 from the final refined class of particles. (F) FSC calculated for the refined model and the final map (black lane). Cross-validation by comparing the FSC calculated against a model refined with a half map (red line) and the FSC calculated with the half map not included in the refinement (blue line). (G) Unsharpened map colored according to the computed local resolution for the final map for the CrPV-IRES and eEF2. (H) Refined model of eEF2 coloured according to the atomic B-factor values (blue low, red high).
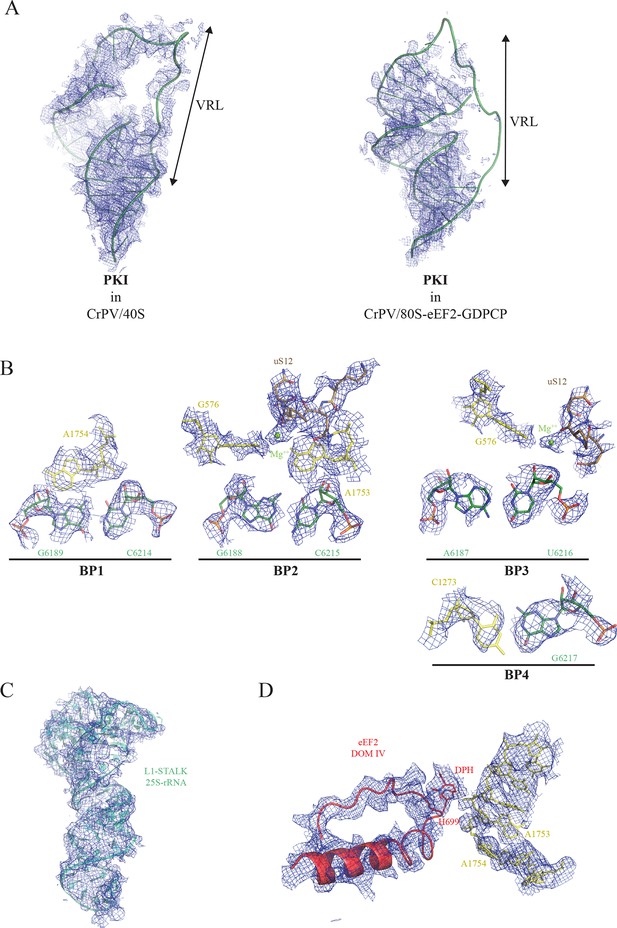
Experimental electron density examples.
(A) CrPV-IRES PKI densities obtained for the CrPV-IRES/40S complex (left) and for the 80S/CrPV-IRES/eEF2 complex (right). Partial density was observed for the variable loop of PKI (VRL) in the complex with the 40S ribosomal subunit. (B) Experimental density observed for the three decoding base pairs of the CrPV-IRES (BP1-3) and the adjacent base pair (BP4) in the 40S/CrPV-IRES complex. CrPV-IRES bases are depicted green and the 40S components of the decoding center yellow. (C) Electron density observed for the rRNA component of the L1-stalk in the displaced conformation as observed in the 80S/CrPV-IRES/eEF2 complex. (D) Electron density for the decoding bases A1753 and 1754 in a retracted position in the 80S/CrPV-IRES/eEF2 complex.
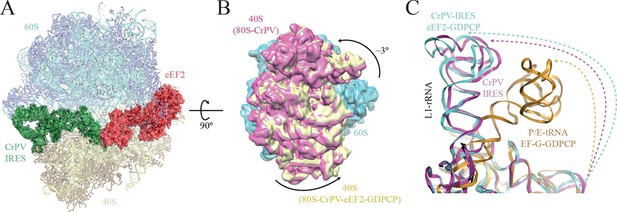
Conformational changes in the complex of CrPV-IRES with the 80S ribosome and eEF2.
(A) Overview of the complex with the large ribosomal subunit (60S) depicted in blue, the small ribosomal subunit (40S) yellow, the CrPV-IRES green and the eEF2 in red. CrPV-IRES and eEF2 are represented as cartoon models inside the corresponding semi-transparent computed molecular surfaces. (B) Superposition of low-pass filtered (8Å) volumes from a front view of the 40S showing an additional ~3º rotation induced in the small subunit by eEF2 binding (yellow) when compared with the rotated state without eEF2 (violet, EMDB-2603). No swivelling of the 40S-head was observed. (C) Additional displacement of the L1 stalk induced by the binding of eEF2. The RNA component of the L1 stalk in the present structure with eEF2 (blue), the rotated state of the complex without eEF2 (violet; PDB 4V92), and in a complex with tRNAs in the hybrid state (orange; PDB-ID 4V9H).
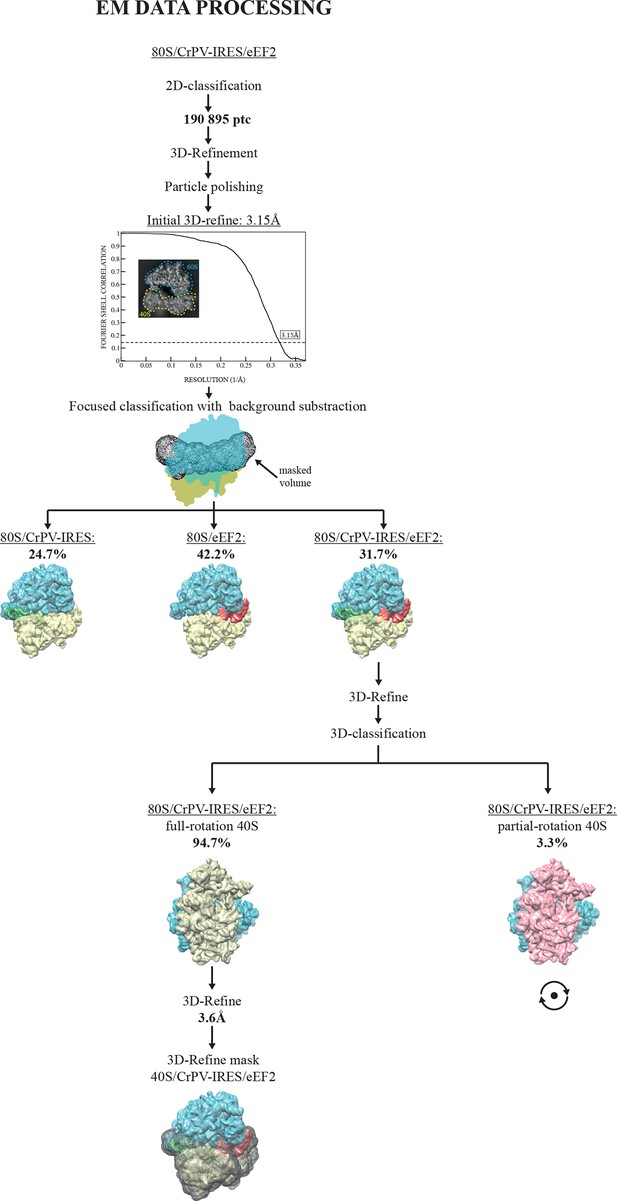
EM image processing workflow for the 80S/CrPV-IRES/eEF2-GDPCP data.
https://doi.org/10.7554/eLife.13567.012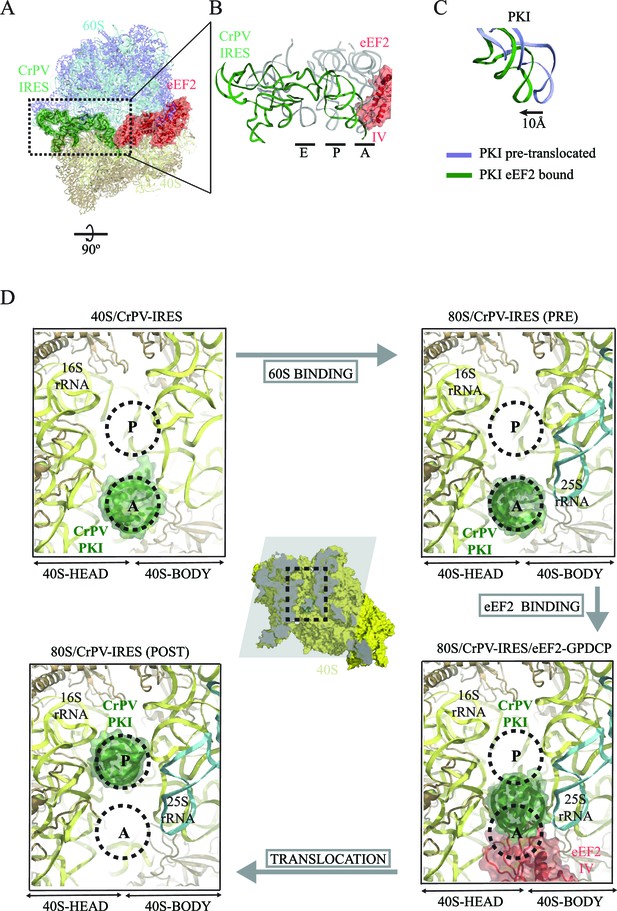
CrPV-IRES conformation induced by the binding of eEF2.
(A) Overview of the complex with the area of interest highlighted. (B) Superposition of the CrPV-IRES (green) with tRNAs in canonical configurations (grey, PDB-ID 2WDG). Ribosomal sites A, P and E are indicated to show how the ASL-like element of the IRES PKI occupies a position between the A and P sites as it interacts with domain IV of eEF2 (red). (C) Relative displacement of PKI in a comparison of the conformation in the present reconstruction (green) with the one adopted in the CrPV-IRES/80S complex (grey). CrPV-IRES superposition has been computed excluding PKI. (D) View down the axis of the PKI stem in the CrPV-IRES/40S complex (top-left), in the pre-translocated CrPV-IRES/80S complex (top-right Fernández et al., 2014), in the 80S/CrPV-IRES/eEF2 complex (bottom-right) and finally, in the post-translocated 80S/CrPV-IRES complex (bottom-left, adapted from Muhs et al. (2015)). Ribosomal sites A and P are depicted as dotted lines. The CrPV-IRES PKI (green) is inserted in the A-site in the isolated interaction with the 40S (yellow) as well as in the pre-translocated 80S/CrPV-IRES complex. The interaction with domain IV of eEF2 (red) displaces it towards the P site so it occupies an intermediate position between the A and P sites.
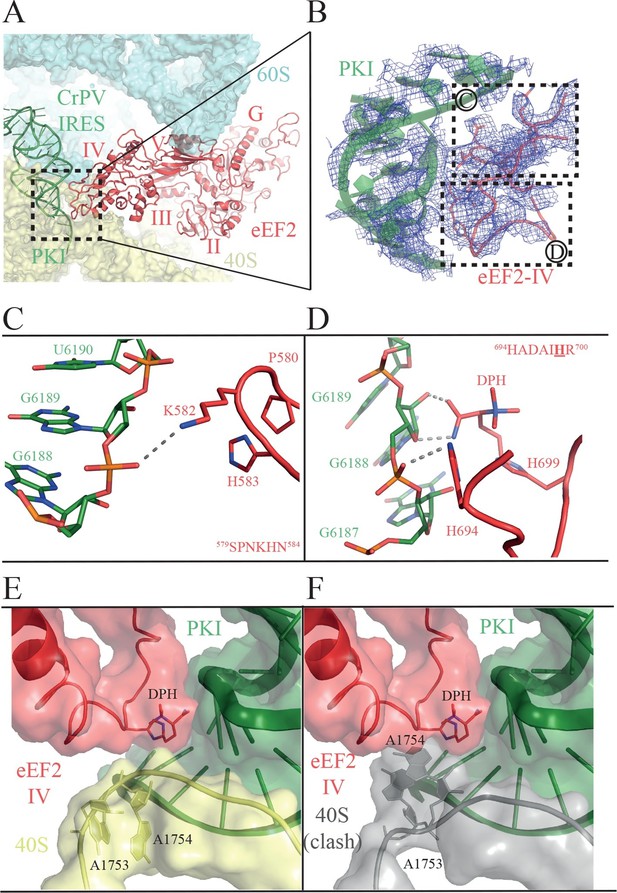
Interactions of PKI of CrPV-IRES with domain IV of eEF2.
(A) Overview of the PKI-eEF2 interaction. The five domains of eEF2 are indicated. (B) Experimental density for the two loops at the tip of domain IV of eEF2 interacting with PKI. (C) Upper loop of eEF2 (residues 579 to 584) implicated in the interaction with PKI. P580 and H583 of eEF2 closely pack together through a stacking interaction to position K582 within interacting distance of the phosphate group of G6189 of the CrPV-IRES. (D) Residues 694–700 in the second loop making direct contacts with PKI. The diphthamide modification of H699 packs against the sugar moiety of G6189. Other residues of this loop such as H694 are also involved in direct contacts with PKI. (E) Retracted position adopted by the decoding bases A1753 and A1754 (A1492 and A1493 in E. coli) in the CrPV-IRES-eEF2-80S complex. (F) The extended conformation of these bases in which they are flipped out of helix 44 as would be seen during decoding or aminoglycoside binding would result in a clash with the tip of eEF2. The small ribosomal subunit components are coloured grey.
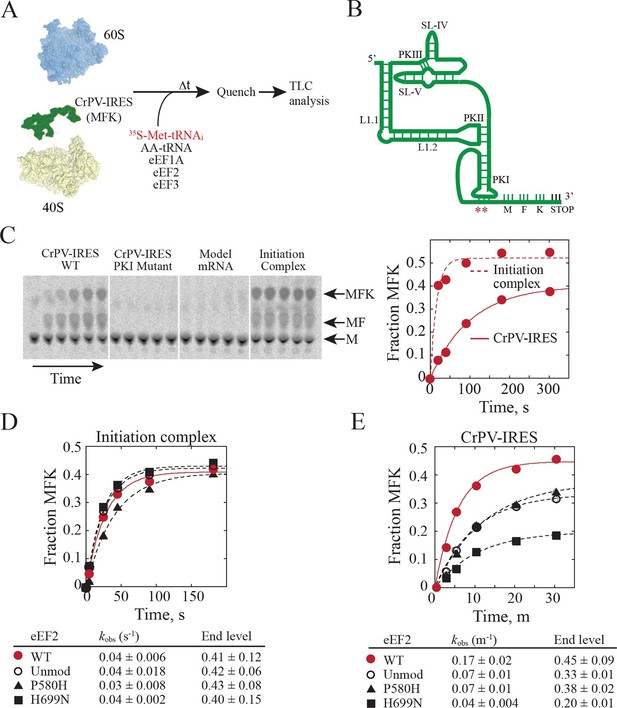
Diphthamide modification on eEF2 promotes CrPV-IRES translation.
(A) Scheme for in vitro reconstituted translation elongation assay with CrPV-IRES RNAs. (B) Diagram of the secondary structure of the 200-nt CrPV-IRES. The coding region of the RNA was altered to encode the tripeptide MFK as indicated. Red asterisks denote the site of the CC to GG mutation in BP1 and BP2 of PKI. (C) Electrophoretic TLC analysis of peptide products from elongation assays programmed with the CrPV-IRES (left panel), the CC to GG mutant CrPV-IRES (second panel), or a model unstructured 52-nt mRNA encoding MFK (third panel). In the fourth panel, the same 52-nt model mRNA was assembled on 80S initiation complexes using the canonical translation initiation factors. The identities of spots corresponding to free methionine (M), dipeptide MF and tripeptide MFK are indicated. The fraction of MFK synthesis in the reactions with the WT CrPV-IRES and the 80S initiation complex were fit to a single exponential equation and plotted on the right. (D–E) Fraction of MFK synthesis in elongation assays programmed with 80S initiation complexes (D) or CrPV-IRES (E) and using purified WT eEF2, unmodified eEF2 lacking diphthamide (Unmod), eEF2-P580H, or eEF2-H699N, as indicated. Results were plotted and fit to a single exponential equation, and observed rate constants and end levels are shown (note the different time scales for the plots). Errors are standard deviation from three independent experiments.
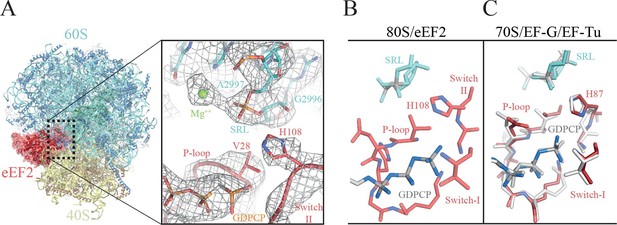
The GTPase center of eEF2 is conserved with those of bacterial EF-G and EF-Tu.
(A) The 80S/CrPV-IRES/eEF2 complex (left) with the inset on the right showing details around the the GTPase centre. Clear experimental density is seen for the GDPCP molecule, the eEF2 residues implicated in catalysis (histidine 108, valine 28), as well as putative magnesium ions (green) that were previously seen with bacterial complexes of GTPase factors. (B) The GTPase centre of eEF2 showing the conformation adopted by the P-loop, switch I and II of the eEF2 as well as the ribosomal elements from the sarcin-ricin loop (SRL, cyan). (C) Corresponding view of EF-G (colored) (Tourigny et al., 2013) and EF-Tu (gray) (Voorhees et al., 2010) bound to the bacterial ribosome.
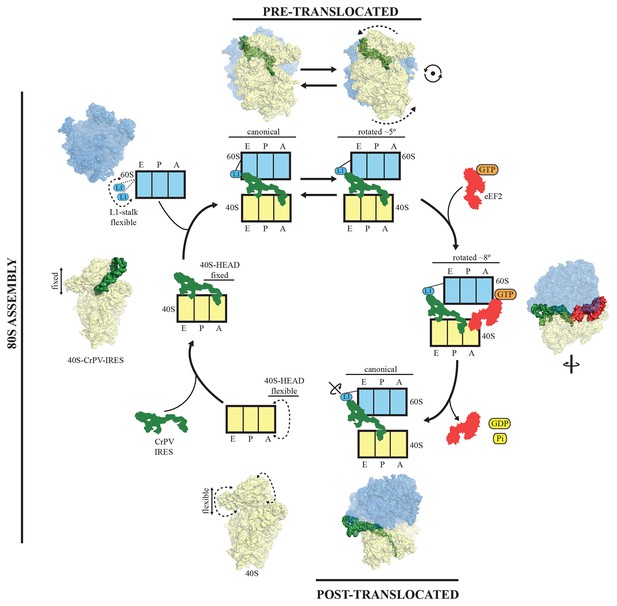
A model for initiation of translation by type IV IRESs.
Starting from the bottom left, type IV IRESs are able to establish a stable binary interaction with the 40S through the IRES elements SL-IV and V. This interaction restricts the flexibility of the 40S-head in a conformation that both allows decoding of PKI and recruitment of the 60S subunit. Top: once the 60S subunit is recruited, the binary CrPV-IRES/80S complex oscillates between canonical and rotated states mimicking the hybrid states of tRNAs (Fernández et al., 2014; Koh et al., 2014). Right: eEF2 in its GTP form binds to the rotated state and induces an extra ~3 degree of rotation of the 40S. This extra rotation results in the IRES interacting with the L1 stalk of the 60S in a wider conformation. Domain IV of eEF2 contacts PKI of the IRES and stabilizes it in an intermediate ap/P-like state. Bottom, post-translocated state (Muhs et al., 2015): GTP hydrolysis results in conformational changes in eEF2 and the ribosome, the translocation of PKI in the P site, breaking of the original contacts of the IRES elements SL-IV and V with the 40S, and release of eEF2. In order for the IRES to adopt such a conformation, a rotation along its longitudinal axes is necessary.
Videos
Video showing the movements of the CrPV-IRES induced by the binding of eEF2 in first translocation event.
CrPV-IRES populates an intermediate state between the A site and the P site reminiscent of a ap/P like state. In order to highlight the movement of CrPV-IRES the motions of the 40S ribosomal subunit in relation to the 60S ribosomal subunit have not been shown.
Video showing experimental density obtained for PKI (green) and the decoding elements of the 40S (yellow) in the CrPV-IRES/40S complex.
https://doi.org/10.7554/eLife.13567.008Video showing experimental density obtained for SL-IV (green) of the CrPV-IRES and ribosomal proteins uS11 (yellow) and uS7 (brown) of the 40S in the CrPV-IRES/40S complex.
https://doi.org/10.7554/eLife.13567.009Video showing experimental density obtained for SL-V (green) of the CrPV-IRES and ribosomal proteins uS7 (yellow) and eS25 (brown) of the 40S in the CrPV-IRES/40S complex.
https://doi.org/10.7554/eLife.13567.010Video showing experimental density obtained for eEF2 (red ribbon) in the 80S/CrPV-IRES/eEF2 complex.
https://doi.org/10.7554/eLife.13567.013Video showing experimental density obtained for PKI (green) and domain IV of eEF2 (red) in the 80S/CrPV-IRES/eEF2 complex.
https://doi.org/10.7554/eLife.13567.016Video showing experimental density obtained for the GTPase Center in the 80S/CrPV-IRES/eEF2 complex.
GDPCP is depicted as grey sticks, eEF2 components are depicted red (switch I as cartoon, switch II as sticks, and P-loop as ribbons) and the sarcin-ricin loop of the 25S rRNA as cyan cartoon. Magnesium ions discussed in the text are represented as green spheres.
Tables
Refinement and model statistics for the final refined models.
| Data Collection | CrPV-IRES/40S | CrPV-IRES/eEF2/80S |
|---|---|---|
| Particles | 54,481 | 37,844 |
| Pixel size (Å) | 1.34 | 1.34 |
| Defocus range (µm) | 1.8-3.5 | 1.8–3.5 |
| Voltage (kV) | 300 | 300 |
| Electron dose (e-/Å2) | 25 | 31 |
| Model composition | ||
| Non-hydrogen atoms | 79,660 | 205,348 |
| Protein residues | 4,828 | 11,278 |
| RNA bases | 1,781 | 5,264 |
| Refinement | ||
| Resolution used for refinement (Å) | 3.8 | 3.6 |
| Map sharpening B-factor (Å2) | -119.8 | -70.8 |
| Average B factor (Å2) | 152.3 | 141.16 |
| MolProbity Clashscore | 97th percentile | 97th percentile |
| MolProbity Score | 90th percentile | 90th percentile |
| Average Fourier Shell Correlation (FSCavg) | 0.72 | 0.83 |
| Rms deviations | ||
| Bonds (Å) | 0.0057 | 0.0072 |
| Angles (˚) | 1.21 | 1.3 |
| Ramachandran plot | ||
| Favored (%) | 84.17 | 81.64 |
| Outliers (%) | 4.4 | 4.63 |
| RNA validation | ||
| correct sugar puckers (%) | 94.2 | 95.3 |
| good backbone conformations (%) | 55.5 | 65.1 |





