The cell proliferation antigen Ki-67 organises heterochromatin
Figures
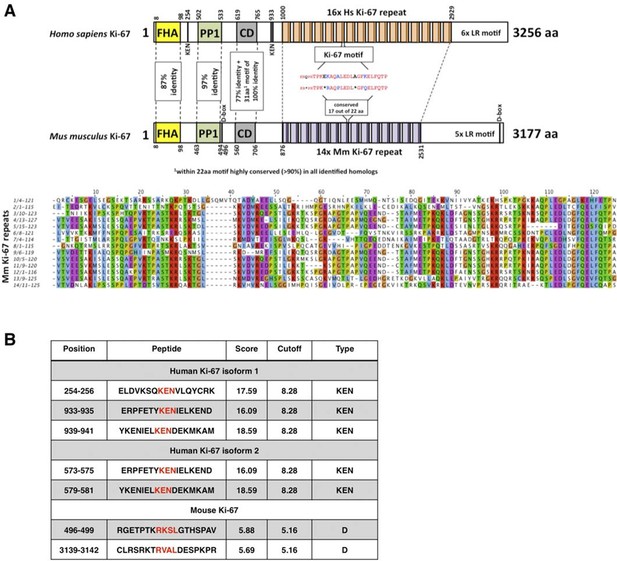
Comparison of human and mouse Ki-67 structural elements.
(A) Top: cartoon of human (long form) and mouse Ki-67 protein highlighting conserved elements and functional motifs. Domains are indicated by boxes (FHA, forkhead-associated domain; PP1, PP1-binding domain; CD, conserved domain; D-box: APC/C targeting destruction box motifs; KEN: APC/C-Cdh1 targeting KEN box motifs). Highly conserved regions are indicated by dotted line with percent of identical amino acids. Bottom: alignment of mouse Ki-67 repeats. (B) APC/C targeting motifs identified in human (both isoforms) and mouse Ki-67.
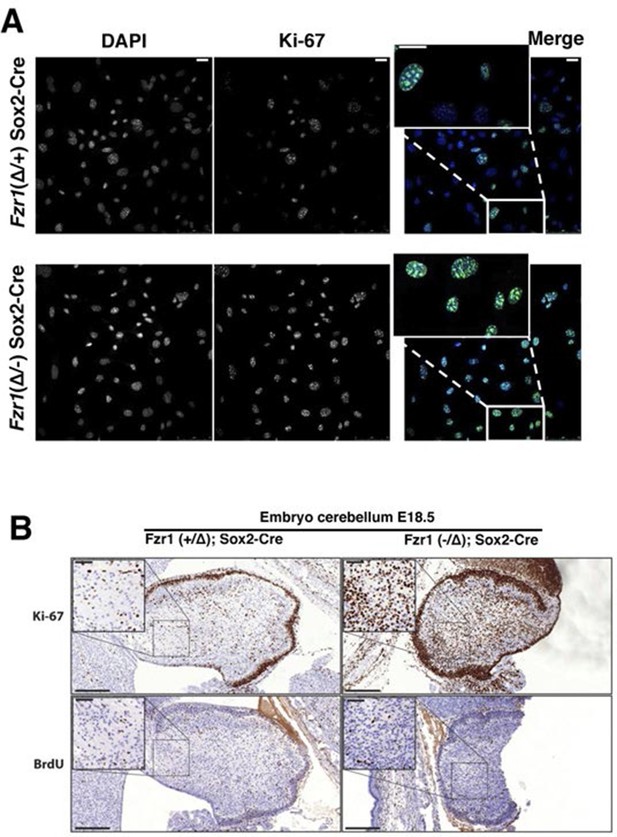
Maintenance of Ki-67 expression in quiescent cells in vivo by Cdh1 mutation.
(A) Immunofluorescence analysis of Ki-67 in MEF cells isolated from embryo (E13.5) of Fzr1(+/Δ);Sox2-Cre and Fzr1(-/Δ);Sox2-Cre mice. Scale bar, 25 µm. (B) IHC staining of Ki-67 and BrdU in sagittal sections of embryo cerebellum (E18.5) of Fzr1 (+/Δ);Sox2-Cre and Fzr1 (-/Δ);Sox2-Cre mice. Bars, 200 µm. Bars in zoom, 50 µm.
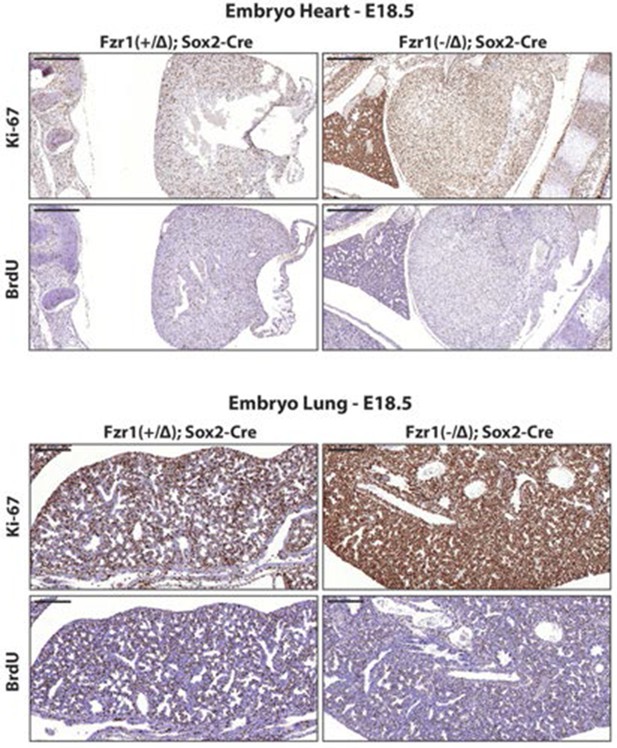
Ki-67 expression is restricted to proliferating cells by APC/C-Cdh1.
Top, IHC staining of Ki-67 and BrdU in sagittal section of embryo heart (E18.5) of Fzr1(+/Δ);Sox2-Cre and Fzr1(-/Δ);Sox2-Cre mice. Scale bar, 500 µm. Bottom, IHC staining of Ki-67 and BrdU in sagittal section of embryo lung (E18.5) of Fzr1(+/Δ);Sox2-Cre and Fzr1(-/Δ);Sox2-Cre mice. Scale bar, 200 µm.
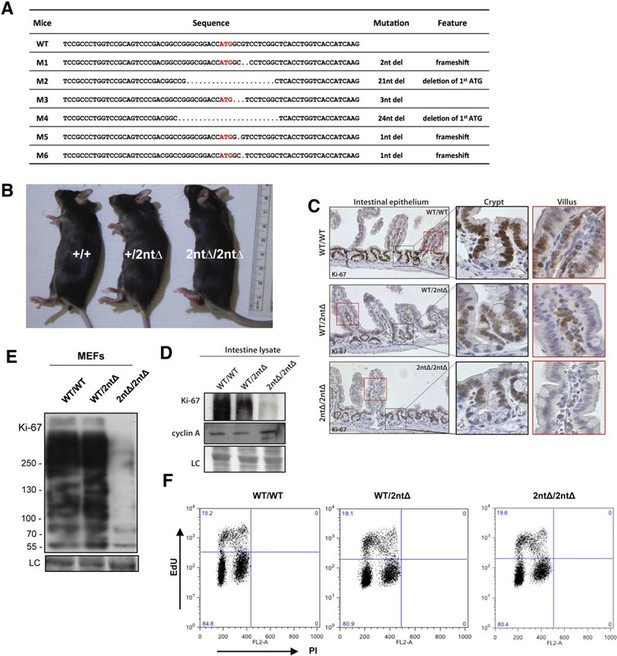
Mouse development with a mutated Ki-67 gene.
(A) Table describing Ki-67 mutant mouse lines resulting from germline transmission of mutations generated by cytoplasmic injection of TALEN-encoding mRNA into zygotes. (B) Macroscopic appearance of littermate female mice at 10 weeks of age. Genotypes are specified. (C) IHC staining of Ki-67 in sagittal section of intestine from Mki67WT/WT, Mki67WT/2nt∆ and Mki672nt∆/2nt∆ mice. (D) Western blots of Ki-67 and cyclin A expression from intestine isolated from Mki67WT/WT, Mki67WT/2nt∆ and Mki672nt∆/2nt∆ mice. LC, loading control. (E) Western blot of Ki-67 in MEFs from WT, Mki67WT/2nt∆ and Mki672nt∆/2nt∆ mice. LC, loading control. (F) Flow cytometry profiles in WT, Mki67WT/2nt∆ and Mki672nt∆/2nt∆ MEFs showing EdU incorporation upon a 1 hr pulse and DNA content.
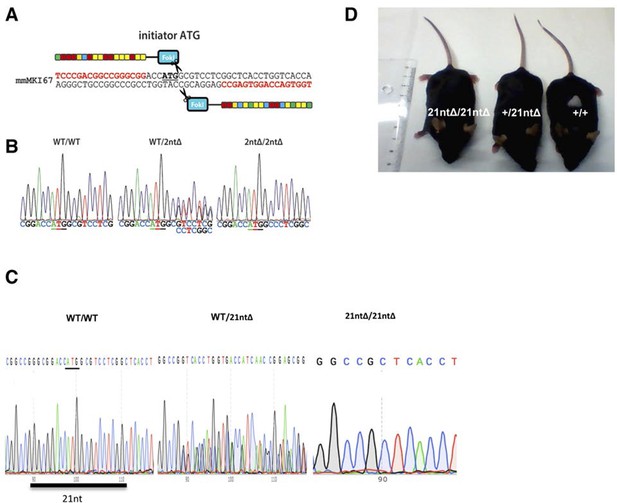
Ki-67 mutant mice develop normally.
(A) Pair of TALE-nucleases designed to target the initiator ATG of mouse Mki67 gene. (B) Sequencing traces of initiator ATG (underlined) of Mki67 gene in WT Mki67+/+ (WT/WT), heterozygous Mki67+/2nt∆(WT/2nt∆) and homozygous Mki672nt∆/2nt∆(2nt∆/2nt∆) mice. (C) Sequencing traces of initiator ATG (underlined) of Mki67 gene in WT Mki67+/+ (WT/WT), heterozygous Mki67+/21nt∆(WT/21nt∆) and homozygous Mki6721nt∆/21nt∆(21nt∆/21nt∆) mice. (D) WT Mki67+/+ (+/+), heterozygous Mki67+/21nt∆(+/21nt∆) and homozygous Mki6721nt∆/21nt∆(21nt∆/21nt∆) mice.
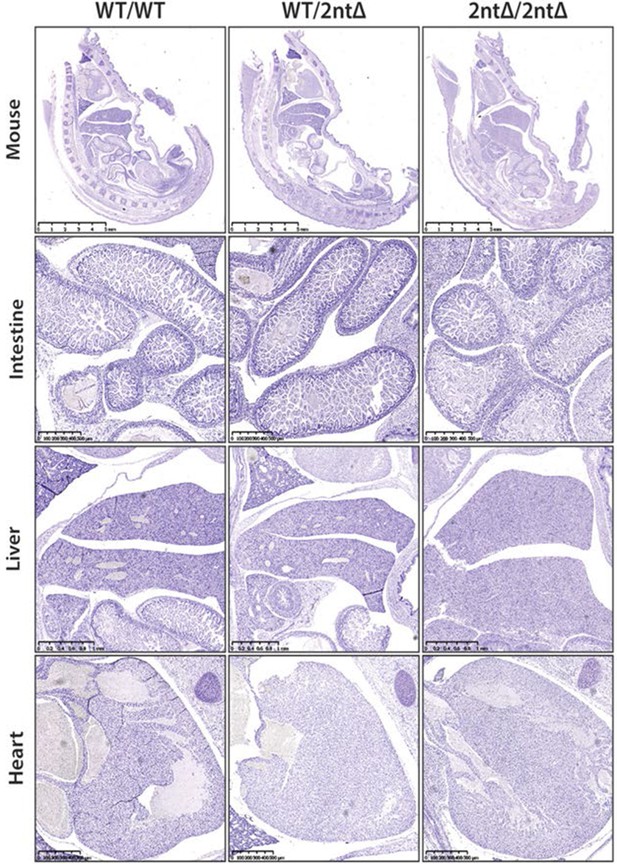
Ki-67 mutant mice develop normally.
Sagittal sections of the whole mouse, intestine, liver and heart from WT Mki67+/+ (WT/WT), heterozygous Mki67+/2nt∆(WT/2nt∆) and homozygous Mki672nt∆/2nt∆(2nt∆/2nt∆) mice. Mouse sections, bar 5 mm; intestine sections, bar 500 µm; liver sections, bar 1 mm; heart sections, bar 500 µm.
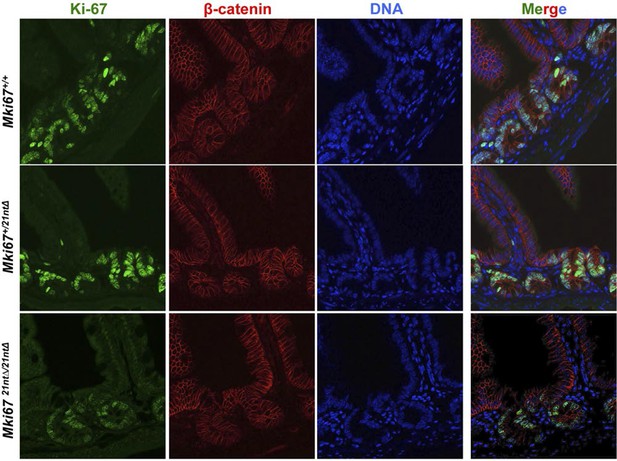
Background Ki-67 levels in Ki-67 mutant mice.
Immunofluorescence of Ki-67 and β-catenin on sagittal sections of the mouse intestinal epithelium from Mki67+/+, heterozygous Mki67+/21nt∆and homozygous Mki6721nt∆/21nt∆mice.
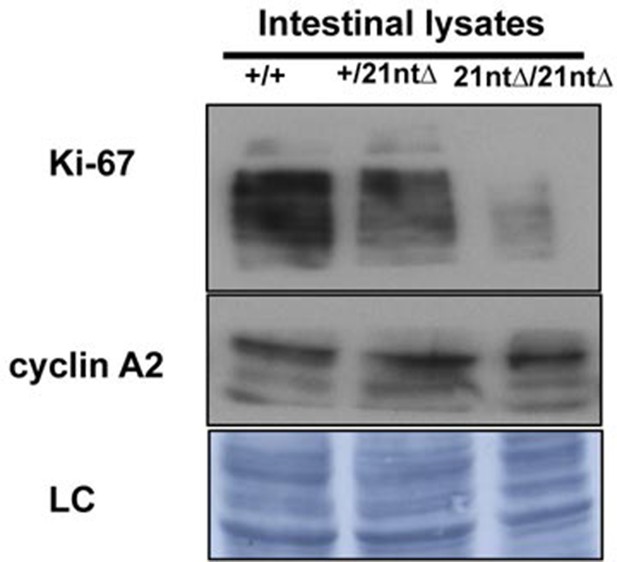
Background Ki-67 levels in Ki-67 mutant mice.
Immunoblotting of Ki-67 and cyclin A on protein preparations of intestinal epithelium from Mki67+/+ (+/+), heterozygous Mki67+/21nt∆(+/21nt∆) and homozygous Mki6721nt∆/21nt∆(21nt∆/21nt∆) mice.
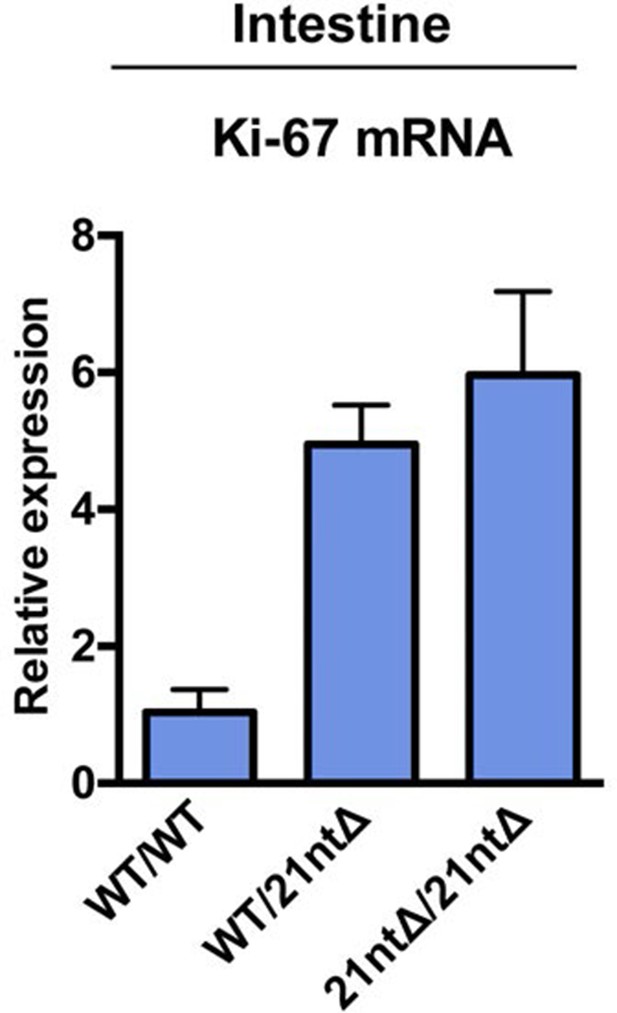
Ki-67 mRNA levels are not reduced in Ki-67 mutant mice.
qRT-PCR of relative Ki-67 mRNA expression, normalised to GAPDH, on preparations of intestinal epithelium from Mki67+/+ (WT/WT), heterozygous Mki67+/21nt∆(WT/21nt∆) and homozygous Mki6721nt∆/21nt∆(21nt∆/21nt∆)mice.
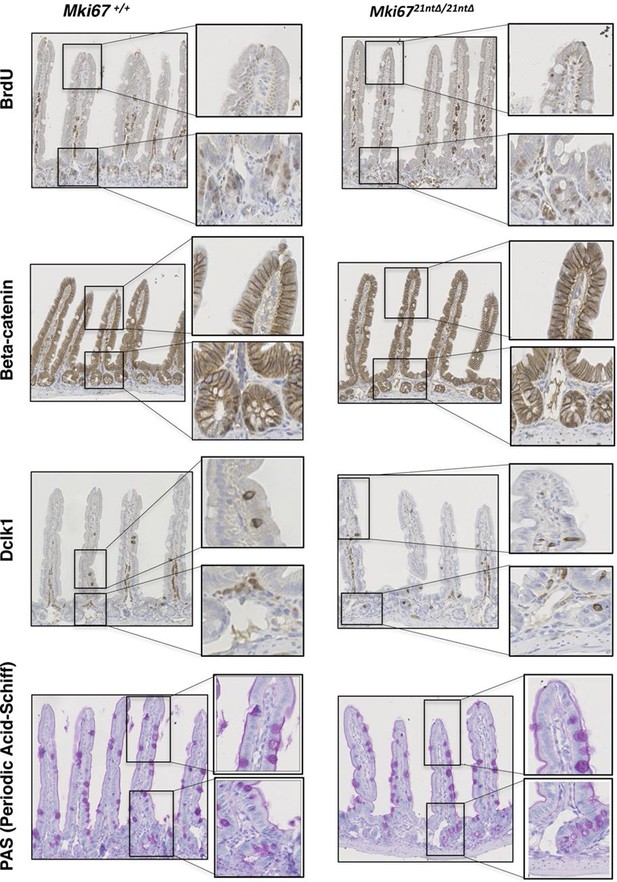
Normal proliferation and differentiation in Ki-67 mutant mice.
Sagittal sections of the intestine from WT Mki67+/+ and homozygous Mki6721nt∆/21nt∆mice sacrificed after a 2 hr BrdU pulse, immunostained for BrdU, β-catenin, DCLK1 (a marker for tuft cells) or PAS staining of mucopolysaccharides (a marker for goblet cells).
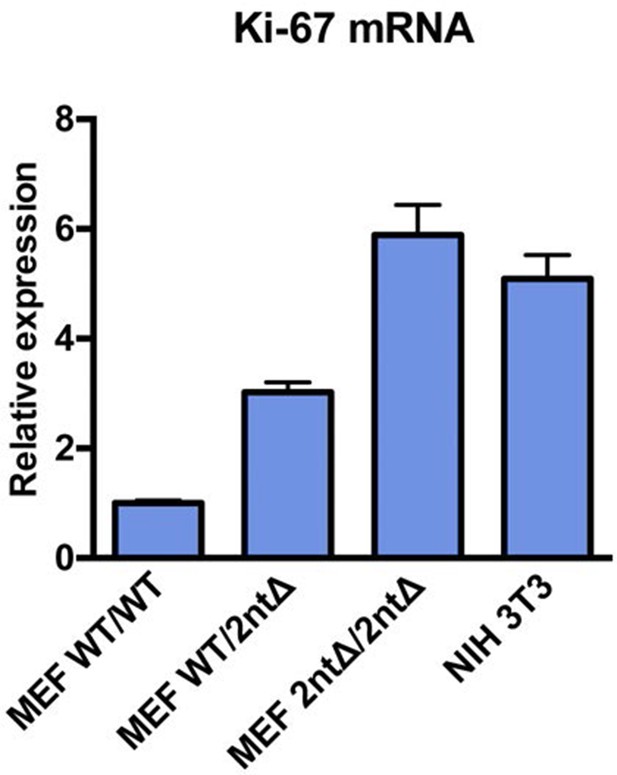
Ki-67 mRNA levels are not reduced in MEFs from Ki-67 mutant mice.
qRT-PCR of relative Ki-67 mRNA expression, normalised to GAPDH, in MEFs isolated from Mki67+/+ (WT/WT), heterozygous Mki67+/2nt∆(WT/2nt∆) and homozygous Mki672nt∆/2nt∆(2nt∆/2nt∆)mice, compared to NIH3T3 cells.
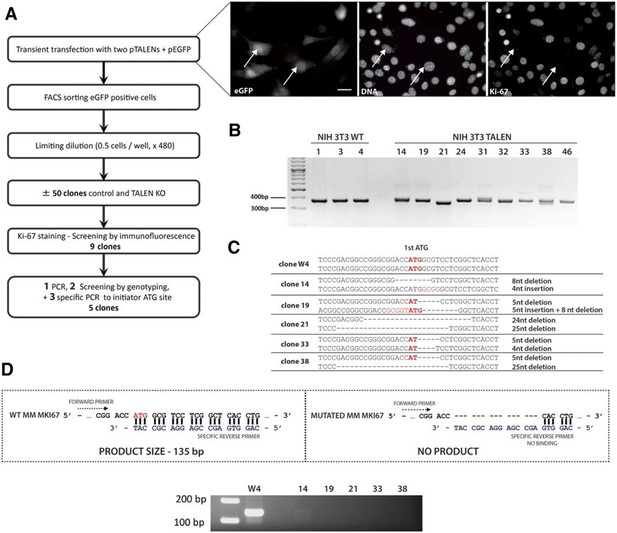
Generation of Ki-67-mutant NIH-3T3 cells.
(A) Top and left, schematic representation of TALEN-mediated generation of NIH-3T3 biallelic Ki-67 mutant cells. Top right, immunofluorescence of Ki-67 and eGFP staining in NIH 3T3 cells transfected with two plasmids encoding TALEN pair and plasmid pEGFP. White arrows show Ki-67-negative pEGFP cotransfected cells. Scale bar 25 µm. (B) PCR analysis of Mki67 initiator ATG surrounding sequence in genomic DNA prepared from three WT clones and nine Ki-67 immunofluorescence-negative clones selected for further analysis. (C) sequencing of Mki67 initiator ATG area from selected clones (14, 19, 21, 33, 38). (D), PCR analysis targeted to the initiator ATG in Mki67 gene of genomic DNA purified from NIH 3T3 WT clone W4 and Ki-67 KO clones 14, 19, 21, 33, 38.
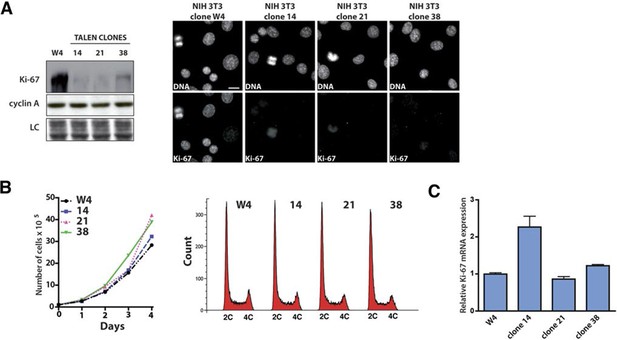
Ki-67-mutant NIH-3T3 cells proliferate normally.
(A) Left, analysis of the indicated protein levels by Western blotting in NIH 3T3 WT clone 4 (W4) and Ki-67 mutant clones 14, 21 and 38. LC, loading control. (B) Left, growth curves of NIH 3T3 WT clone W4 and Ki-67 mutant clones 14, 21 and 38. NIH 3T3 WT and mutant cells were counted every day for 4 days. Right, cell cycle distribution of the WT clone W4 and 14, 21, 38 Ki67 mutant clones as analysed by FACS. (C) qRT-PCR analysis of Ki-67 mRNA in NIH 3T3 WT clone W4 and NIH Ki-67 mutant clones 14, 21 and 38. Normalized by mRNA expression of B2m (beta-2-microglobulin).
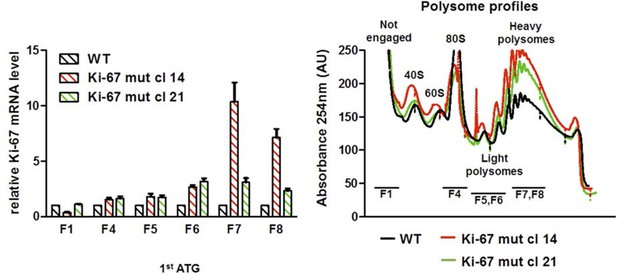
Basal translation of Ki-67 with a mutated ATG.
Left: quantification of association of Ki-67 mRNA with ribosome fractions in biallelic single TALEN Ki-67 mutant NIH-3T3 cells by qRT-PCR, normalised to GAPDH, b2-microglobulin and actin mRNA and to WT levels. Right: RNA quantification of fractions by spectrophotometric absorbance at 254 nm.
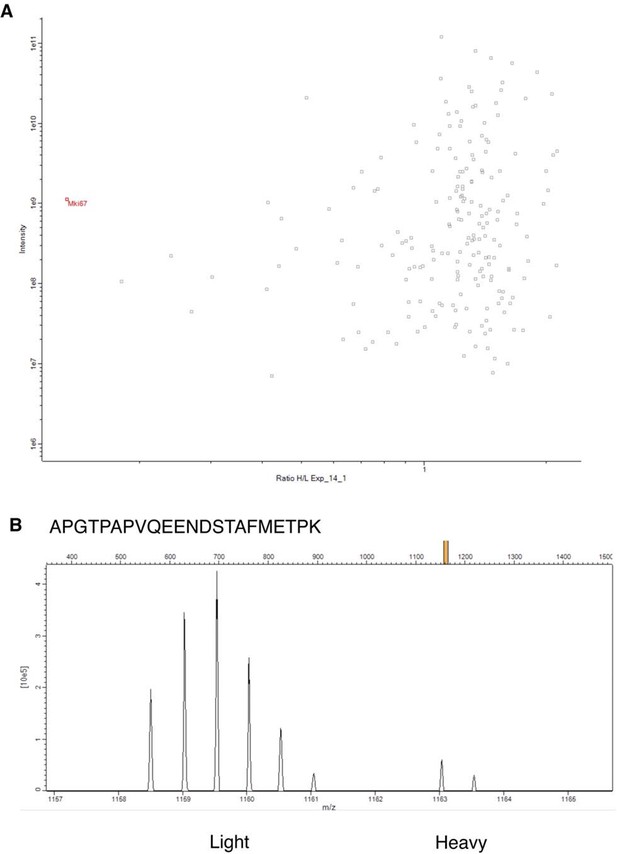
SILAC proteomics analysis of Ki-67 expression with a mutated ATG.
(A) Scatterplot showing H/L m/z peak ratios (x-axis) against intensity levels (y-axis) from band 1 of SDS-PAGE purified chromatin, where H-labelled samples were Ki-67 mutant NIH-3T3 clone 14, L samples were W4 wild-type control. Most proteins have unaltered expression between the two cell lines (H/L ratio ≈ 1). Average H/L ratio of putative Ki-67 peptides (predicted by Maxquant on the basis of isotope profiles with the expected difference in m/z ratio from Ki-67 L-peptides positively identified by MS/MS) is shown in red. (B) Isotope profile of a light Ki-67 peptide from control cell chromatin identified by MS/MS, and the corresponding heavy peaks used for quantification. Data is provided in Figure 3—figure supplement 11—source data 1.
-
Figure 3—figure supplement 11—source data 1
SILAC quantitation of Ki-67 peptides from WT and Mki67-mutant NIH-3T3 cells.
Table showing identity and H/L ratio of Ki-67 peptides purified from chromatin of NIH-3T3 Wt, Mki67 mutant clones 14 and 21, recovered in excised bands 1 (>250 kDa) or 2 (130-250 kDa). Data have been uploaded into the massIVE repository with accession number MSV000079492.
- https://doi.org/10.7554/eLife.13722.018
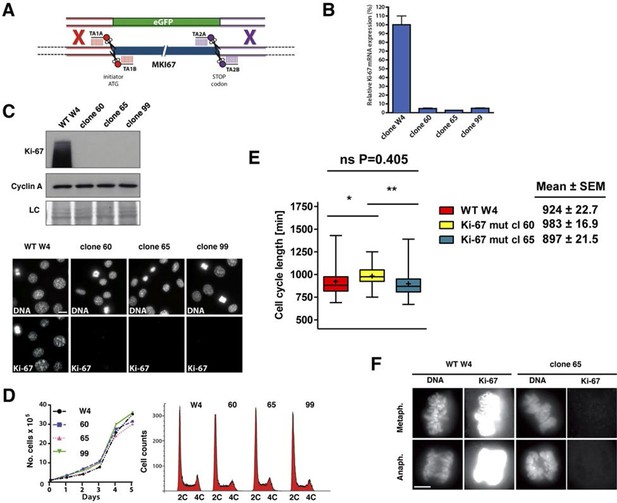
Cell proliferation without Ki-67.
(A) Schematic representation of strategy for TALEN-mediated generation of Mki67 null allele. (B) qRT-PCR analysis of Ki-67 mRNA levels in NIH-3T3 WT clone W4 and Ki-67-negative 60, 65, 99 clones. (C) Top: Western blot of Ki-67 and Cyclin A in NIH-3T3 WT clone W4 and Ki-67-negative mutant clones 60, 65, 99; LC, loading control; below, Ki-67 immunofluorescence; bar, 10 µm. (D) Left, growth curves of WT and Ki-67 null cell lines 60, 65 and 99; right, cell cycle distribution analysed by flow cytometry. (E) Cell cycle length of WT clone W4 and Ki-67 null clones 60 and 65 as determined by time-lapse videomicroscopy. (F) Cells of clone 65 show altered chromosomal periphery in mitosis. The Ki-67 staining is deliberately overexposed to demonstrate absence of detectable Ki-67 in clone 65, even in metaphase. Bar, 5 µm.
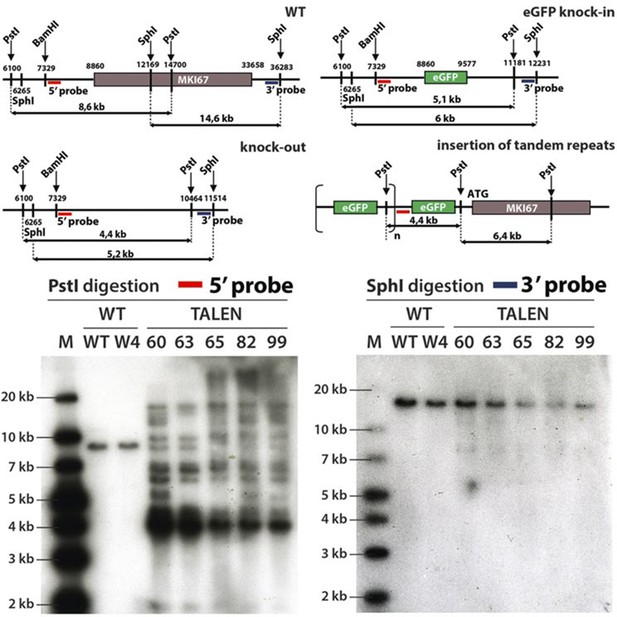
Generation of NIH-3T3 cells lacking Ki-67.
Top, schematic representation of the wild type (WT), knock-out or eGFP knock-in Mki67 locus and the predicted insertion of tandem repeats of the eGFP insert upstream of Mki67 locus. Bottom, Southern-blot of two NIH-3T3 WT clones (WT, W4) and six NIH-3T3 Ki-67 mutant clones (60,63,65,82,99). Clones were digested with PstI or SphI and probed with 5’ or 3’ probe, respectively.
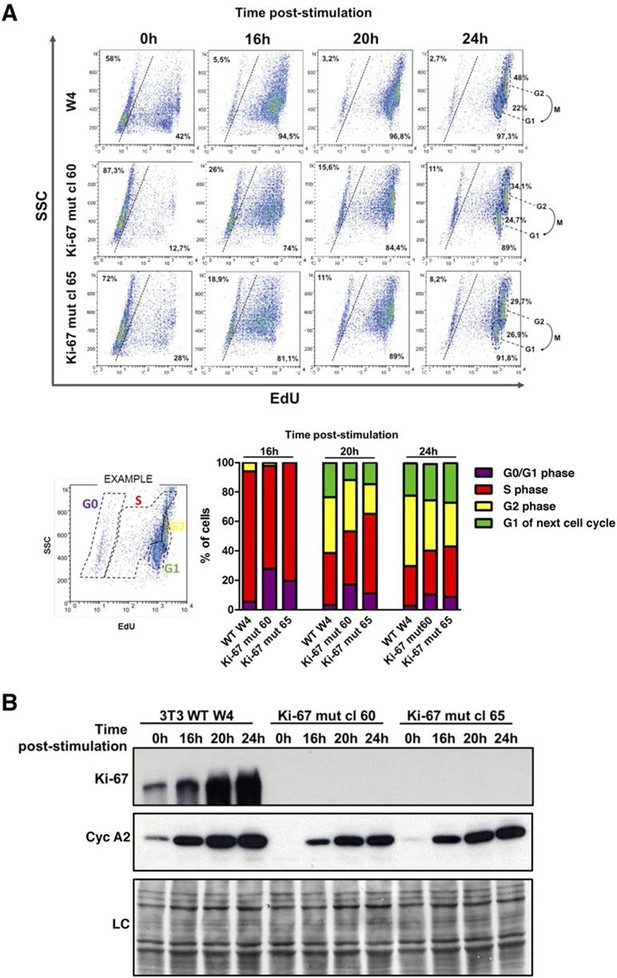
Cells lacking Ki-67 enter the cell cycle efficiently.
(A) Top, re-entry of cell cycle in NIH-3T3 WT clone W4 and Ki-67-negative mutant clones 60 and 65 after serum starvation-induced cell cycle arrest. Progression of cell cycle entry analysed by FACS using EdU staining. Bottom, quantification of cell cycle phases in this experiment. (B) Western blot analysis of Ki-67 (upper panel) and cyclin A2 (lower panel) upon cell cycle entry. LC, loading control.
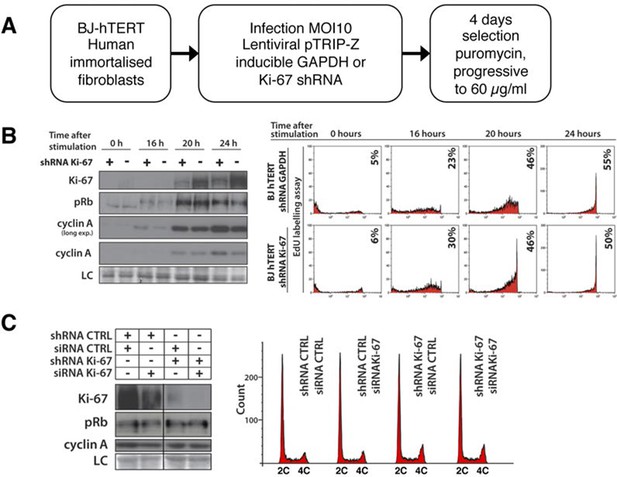
Cells lacking Ki-67 proliferate efficiently.
(A) Schematic presentation of generation of Ki-67 shRNA knockdown BJ-hTERT. (B) Left, Western blot for indicated proteins upon cell cycle re-entry in serum starved hTERT-transformed BJ fibroblasts (BJ-hTERT) after induction of shRNA against Ki-67 (+) or GAPDH control (-). LC, loading control. Right, DNA synthesis analysed by flow cytometry after EdU pulse. (C) Left, asynchronous BJ-hTERT with doxycyclin-induced control or Ki-67 shRNA-expression were additionally transfected with control or Ki-67 siRNA for 48 hr. Protein levels were analysed by Western blotting. LC, loading control. Right, cell cycle distribution by flow cytometry.
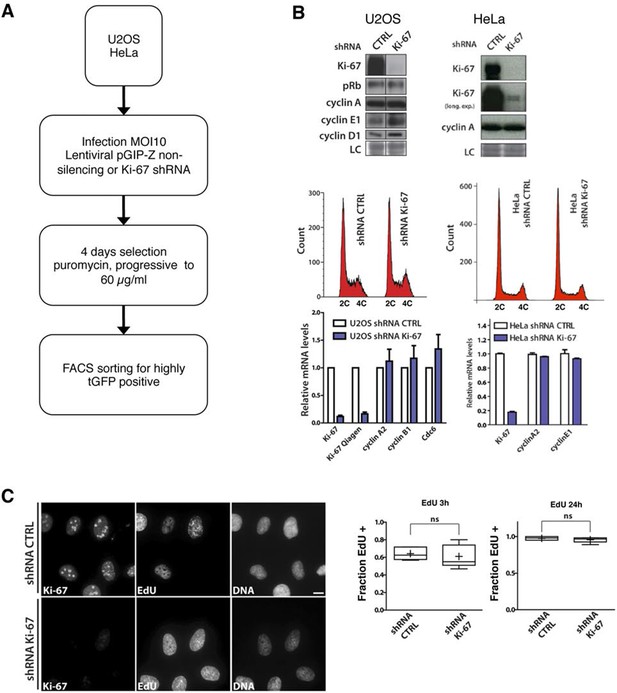
Cells lacking Ki-67 proliferate efficiently.
(A) Schematic presentation of generation of Ki-67 shRNA knockdown cell lines. (B) Top: Western blot analysis of the indicated proteins in asynchronously growing U2OS and HeLa cells stably expressing non-targeting (CTRL) or Ki-67 shRNA. Lanes separated by lines were from a single exposure of a single SDS-PAGE gel and Western blot. LC, loading control. Middle: Cell cycle distribution of these cells. Bottom: qRT-PCR analysis of the indicated mRNA levels, normalized by mRNA expression of beta-2-microglobulin (B2m). (C) Immunofluorescence of Ki-67 and EdU in asynchronous U2OS cells stably expressing control or Ki-67 shRNA, incubated with 5-ethynyl-2’-deoxyuridine (EdU) for 3 hr. Bar, 10 µm. Right: Ratio of EdU-positive cells to the total cell number for each incubation time; ns: not significant.
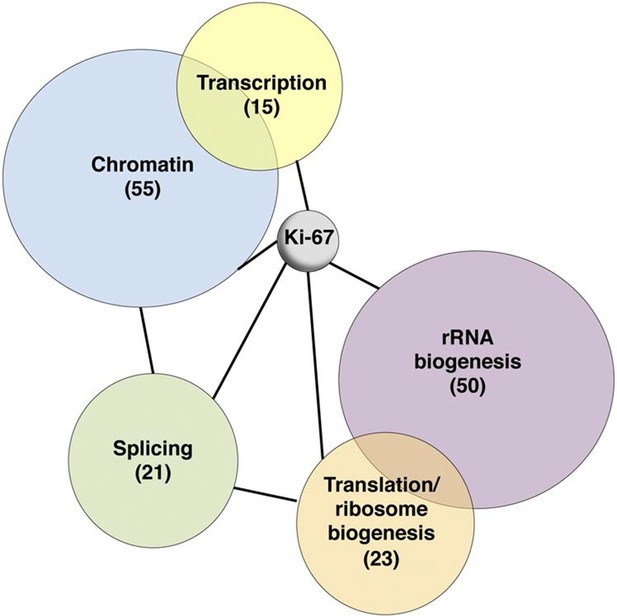
Ki-67 interacts with proteins involved in nucleolar processes and chromatin.
Simplified STRING analysis reveals network interactions between proteins associating with Ki-67. The full network is shown in Figure 6—figure supplement 2; data is provided in Figure 6—source data 1.
-
Figure 6—source data 1
Ki-67 interacting proteome.
Table showing proteins identified by mass spectrometry eluted from immunoprecipitates of cells transfected with FLAG-tagged Ki-67, an unrelated protein (FLAG-tagged TRIM39) or empty vector. Proteins specifically interacting only with Ki-67 are presented on the first worksheet. Data are available via ProteomeXchange with identifier PXD003551.
- https://doi.org/10.7554/eLife.13722.025
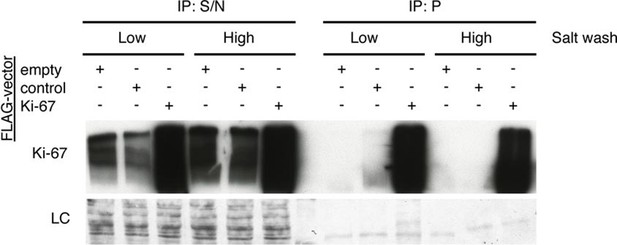
The Ki-67 interactome.
Western blot of Ki-67 in nuclear extracts from cells expressing FLAG-tagged versions of Ki-67 or a control unrelated protein, or FLAG alone. IP: immunoprecipitation; S/N: supernatant; P: pellet. Bottom, loading control.
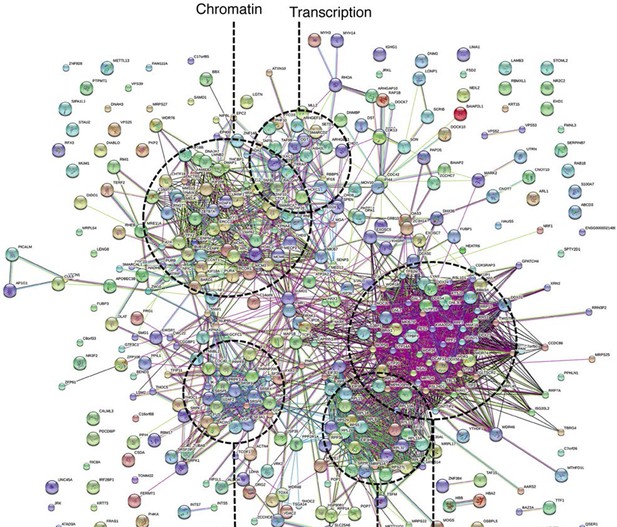
Ki-67 interacts with proteins involved in nucleolar processes and chromatin.
STRING analysis of network interactions between proteins specifically associating with Ki-67 as identified by immunoprecipitation and mass spectrometry from U2OS cells expressing FLAG-tagged full length human Ki-67.
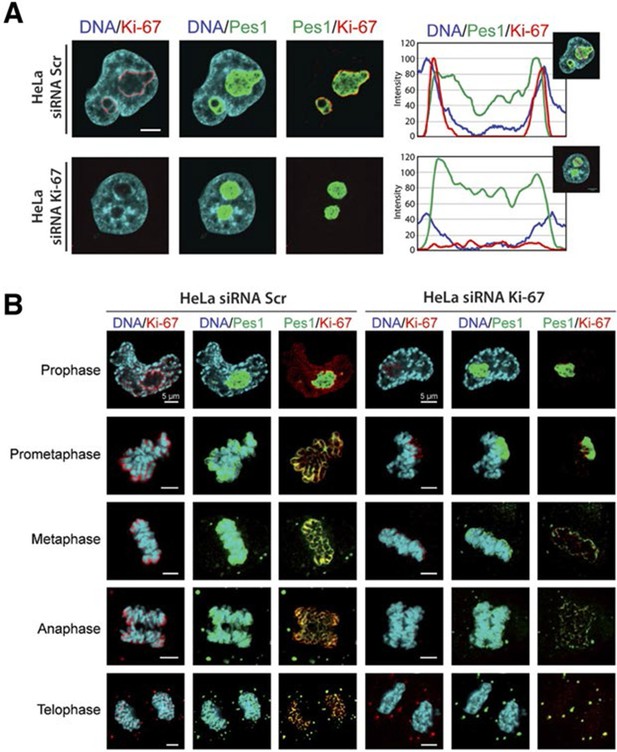
Ki-67 localises PES1 to mitotic chromosomes.
(A) Analysis of the interphase localisation of PES1 and Ki-67 proteins by immunofluorescence in HeLa cells 72 hr after transfection with control siRNA (scramble; Scr) or Ki-67 RNAi. Right, line scans showing the distribution of fluorescence signals within indicated nucleoli (dashed line). Images were captured in confocal mode with a spinning-disk microscope. Bar, 5 μm. (B) Analysis of the mitotic localisation of PES1 and Ki-67 proteins by immunofluorescence in HeLa cells 72 hr after transfection with control siRNA (scramble; Scr) or Ki-67 RNAi. Bar, 5 μm.
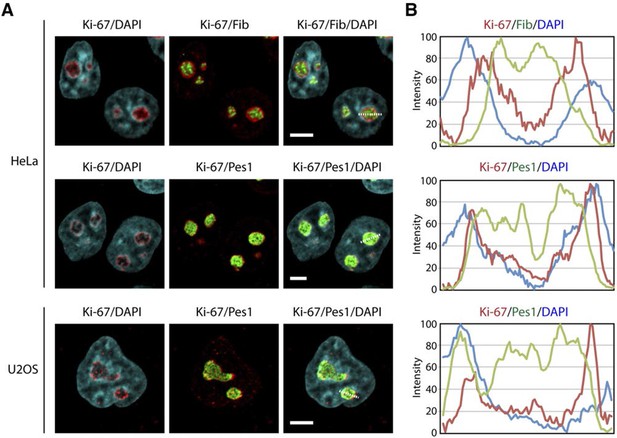
Ki-67 is a nucleolar protein localizing in the cortical side of the GC.
(A) Localisation of Ki-67 protein by immunofluorescence in HeLa and U2OS cells. Fibrillarin (DFC) or Pes1 (GC) were used as nucleolar markers. Images were captured in confocal mode with a spinning-disk microscope. Objective 100 x. Scale bar: 5 μm. (B) Line scans showing the distribution of fluorescence signals within specific nucleoli (see dotted lines in panel A).
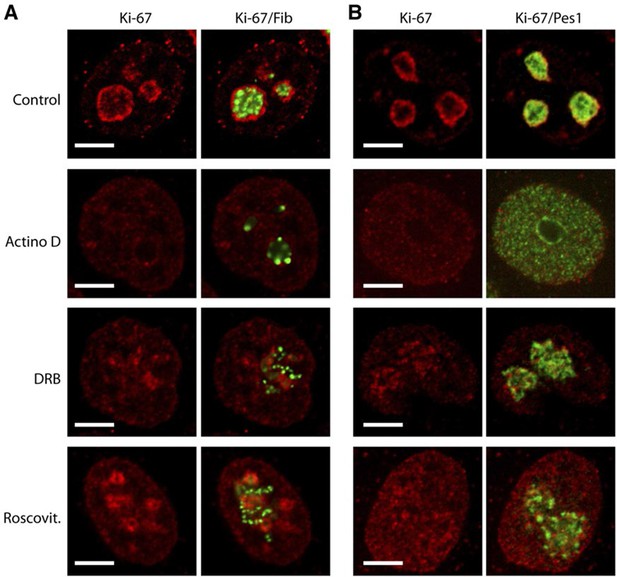
Ki-67 follows GC components upon drug-induced nucleolar disruption.
Actinomycin D or the kinase inhibitors DRB and Roscovitine were added to cells during 90 min and Ki-67 was located within these cells by immunofluorescence. Fibrillarin (A) and PES1 (B) proteins were also localised by immunofluorescence. Objective 100 x. Scale bar: 5 μm.
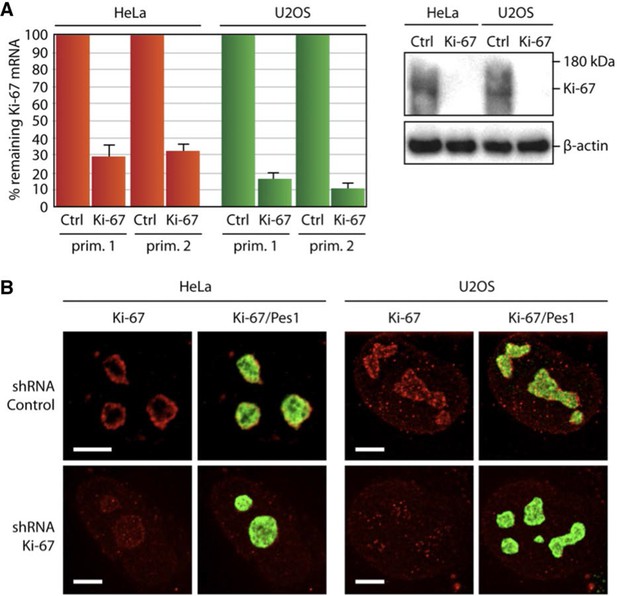
Depletion of Ki-67 does not affect overall nucleolar structure.
(A) Histograms showing the level of Ki-67 mRNA remaining in the cell lines expressing constitutively the shRNA against Ki-67. These levels were assessed by RT-qPCR using two different pairs of primers. Right, the Western-blot against Ki-67 shows a disappearance of a bands in the cell lines constitutively expressing the shRNA against Ki-67. (B) Immuno-fluorescence against Ki-67 in control HeLa and U20S cell lines in the absence (control) or in the presence (Ki-67) of the shRNA targeting Ki-67 mRNA. The PES1 signal shows that the nucleolar structure is maintained. Objective 100 x. Scale bar: 5 μm.
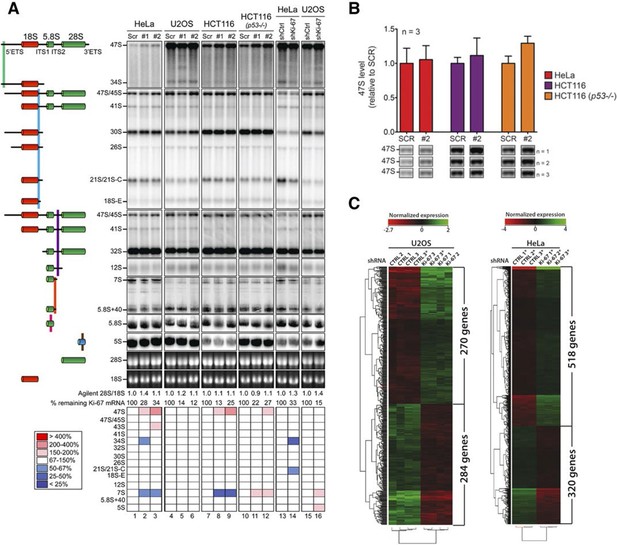
Ki-67 is not required for rRNA biogenesis but controls gene transcription.
(A) Northern-blot analysis of total RNA extracted from HeLa and U2OS cells constitutively expressing shRNA against Ki-67; and HeLa, U2OS, HCT-116 and HCT-116 TP53 (-/-) depleted of Ki-67 by siRNA for 72 hr in two biological replicates (#1 and #2) or with scrambled siRNA control (Scr). Pre-rRNA intermediates were analysed by probing with different primers located in the different spacers of the 47S sequence (5’ETS-green; ITS1-blue; ITS2-purple). (B) Quantification of 47S rRNA precursor in HeLa, HCT-116 and HCT-116 TP53 (-/-) depleted of Ki-67 by siRNA for 72 hr in three biological replicates (n=1–3). (C) U2OS cells (left) or HeLa cells (right) show transcriptome profile differences (fold change >1.5; corrected p-value <0.02) between asynchronous cells constitutively expressing control (CTRL) or Ki-67 shRNA. Heat-maps present the expression levels of differentially expressed genes between biological replicates (1,2,3) and technical replicates (3, 3*). Data is provided in Figure 8—source data 1 and 2.
-
Figure 8—source data 1
Ki-67-dependent transcriptome in U2OS cells.
Table showing statistically significant (corrected p value < 0.02, Fold-change >1.5) changes of transcript abundance from Agilent Gene chip analysis of cDNA from control U2OS (pGIPZ-shRNA non silencing control) and U2OS stably silenced (pGIPZ-Ki-67 shRNA) for Ki-67.
- https://doi.org/10.7554/eLife.13722.033
-
Figure 8—source data 2
Ki-67-dependent transcriptome in HeLa cells.
Table showing statistically significant (corrected p value <0.02, Fold-change >1.5) changes of transcript abundance from Agilent Gene chip analysis of cDNA from control HeLa (pGIPZ-shRNA non silencing control) and HeLa stably silenced (pGIPZ-Ki-67 shRNA) for Ki-67.
- https://doi.org/10.7554/eLife.13722.034
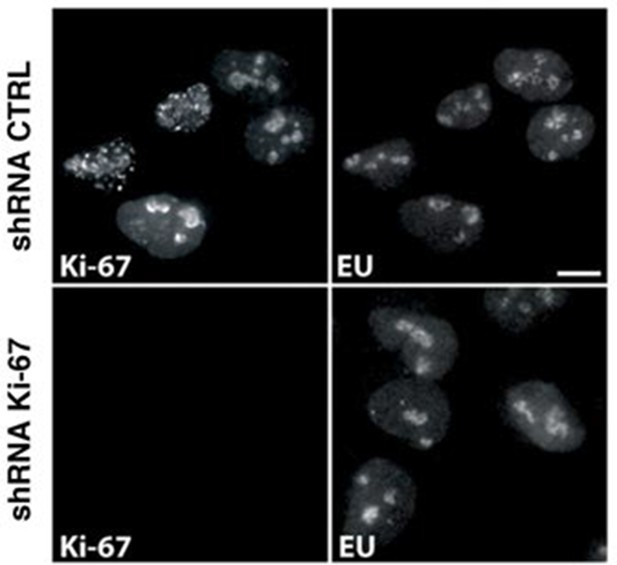
Ki-67 depletion does not hinder rRNA transcription.
Newly synthesised RNA in asynchronous U2OS cells expressing control or Ki-67 shRNA visualised by 5-ethynyl uridine (EU) incorporation and immunofluorescence. Bar, 10 µm.
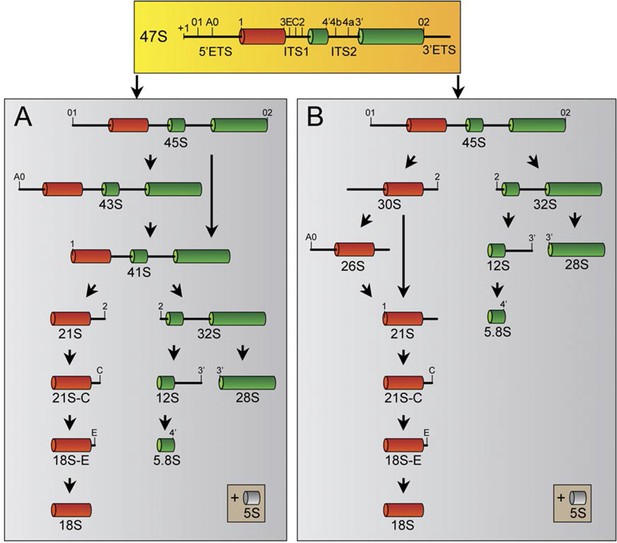
Human pre-rRNA processing pathway involves two major pathways.
The two pathways (A and B) are characterized by specific cleavage kinetics. In pathway A, cleavages at the site A0 and 1 occur before cleavage at the site 2, whereas cleavage at site 2 occurs before cleavages at sites A0 and 1 in the pathway B. Both pathways will end by the production of the mature 28S, 18S and 5.8S. The 5S rRNA is synthesized by the RNA pol III in the nucleoplasm and will join the other rRNAs during ribosome assembly.
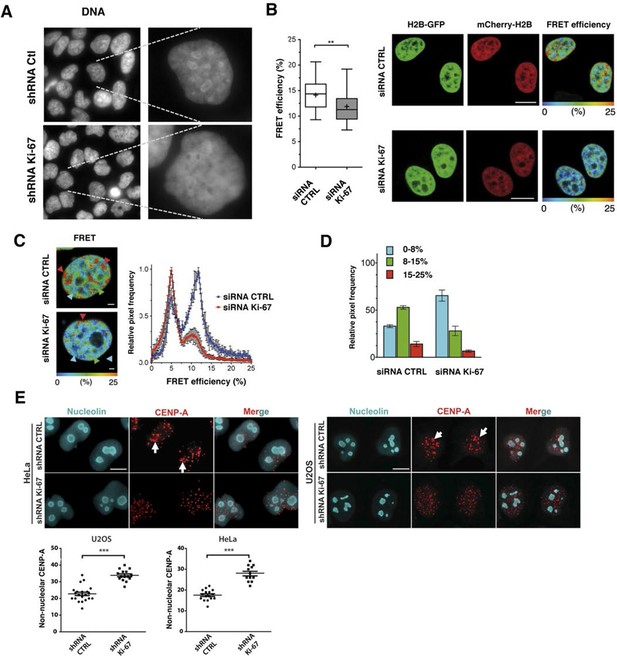
Ki-67 promotes heterochromatin interactions.
(A) DAPI staining in control and stable Ki-67-knockdown U2OS cells. (B) HeLa cells stably expressing GFP-H2B and mCherry-H2B, depleted using Ki-67 or non-targeting (CTRL) siRNA. Left, FRET efficiency (cross shows mean value) ** Different, p<0.01. FRET efficiency and spatial distribution shown by a pseudocolour scale of FRET (%) values from 0 to 25%. Bars, 10 μm. (C) Left, representative HeLaH2B-2FP nuclei showing spatial distribution of FRET efficiency. Arrowheads show different chromatin compaction states (high FRET, red; intermediate, green; low, blue), Bars, 2 μm. Right, mean FRET distribution curves from siRNA control (blue curve, n=8) and siRNA Ki-67 (red curve, n=11) nuclei. (D) Relative fraction of the three FRET efficiency populations (blue (low), FRET efficiency ≤ 8%; green (medium), 8–15%; and red (high), 15–25%) in siRNA control and siRNA Ki-67 nuclei. Error bars indicate SD. (E) Immunofluorescence of CENP-A localisation in control and stable Ki-67 knockdown HeLa (left) and U2OS (right) cells. Nucleolar localisation (white arrows). Nucleolin was used as nucleolar marker. Bar, 10 µm. Below: quantification in different cells of numbers of CENP-A spots not associated with the nucleolus.
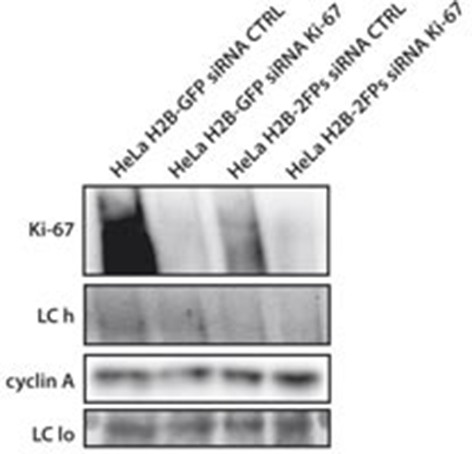
Knockdown of Ki-67 in H2B FRET cell line.
Western blot analysis of the indicated proteins in asynchronously growing HeLa H2B FRET cells transiently transfected with control siRNA (Ctrl) or Ki-67 RNAi for 72 hr. LC, loading controls of the high (h) and low (lo) MW parts of the SDS-PAGE gel.
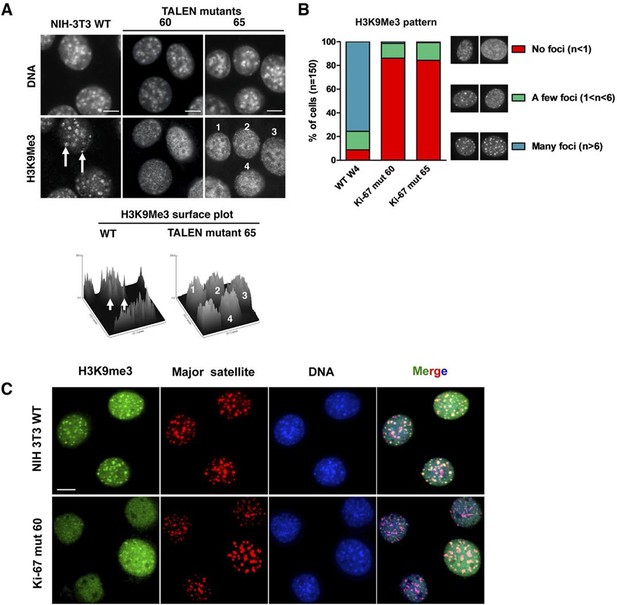
Ki-67 controls heterochromatin organisation.
(A) Top, immunofluorescence analysis of H3K9me3 in mouse NIH-3T3 WT and Mki67 TALEN mutant clones 60 and 65. Bars, 5 µm. Below: graphs showing quantification of pixel intensity scans for H3K9me3. (B) Quantification of H3K9Me3 patterns in WT and Ki-67 mutant clones. (C) Immunofluorescence of H3K9Me3, FISH of major satellite DNA and DAPI staining in WT W4 and Ki-67 mutant clone 60. Bar, 10 µm.
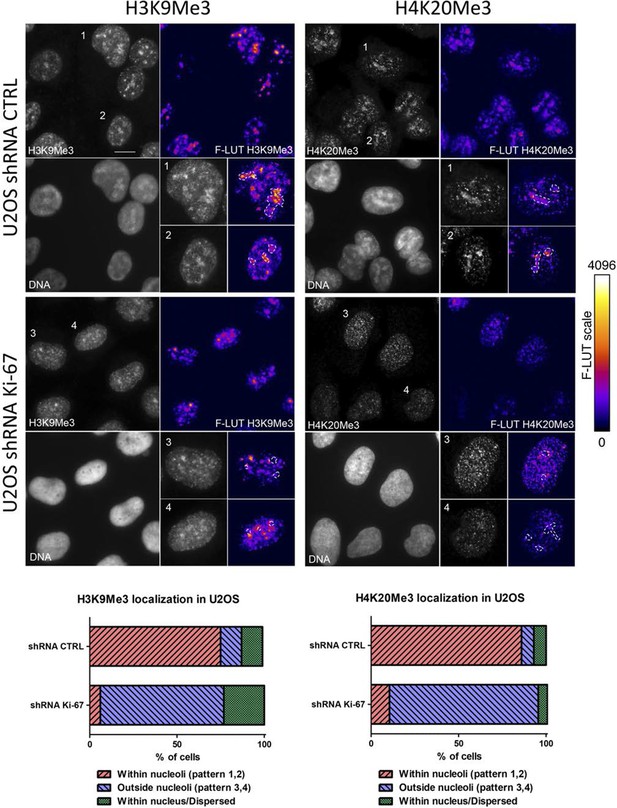
Heterochromatic histone mark localisation requires Ki-67.
Immunofluorescence analysis of H3K9me3 (left) and H4K20me3 (right) localisation in stable control and Ki-67 knockdown U2OS cells. Right: Fire look up table (F-LUT) pseudocolouring of immunofluorescence staining intensity, generated using Fiji software (Schindelin et al., 2012). Dotted white lines denote nucleolus, while numbers 1–4 identify cells for insets as well as staining patterns within nucleolus (1,2) or outside the nucleolus (3,4). Histograms below show the percentage of cells counted showing each pattern. The 2D Fire-LUT surface plot was generated using Fiji software(1). Scale bar: 10 μm.
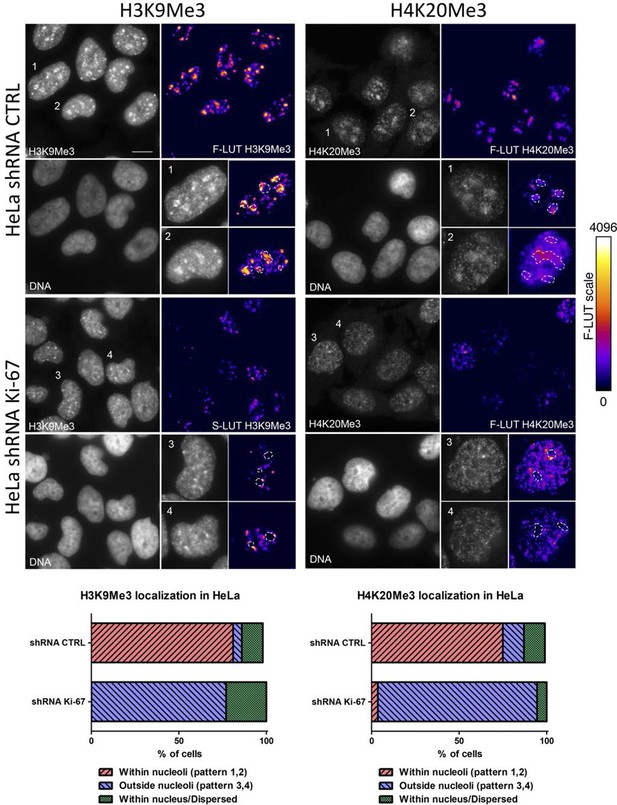
Heterochromatic histone mark localisation requires Ki-67.
Immunofluorescence analysis of H3K9me3 (left) and H4K20me3 (right) localisation in stable control and Ki-67 knockdown HeLa cells. Right: Fire look up table (F-LUT) pseudocolouring of immunofluorescence staining intensity. Dotted white lines denote nucleolus, while numbers 1–4 identify cells for insets as well as staining patterns within nucleolus (1,2) or outside the nucleolus (3,4). Histograms below show the percentage of cells counted showing each pattern. Scale bar: 10 μm.
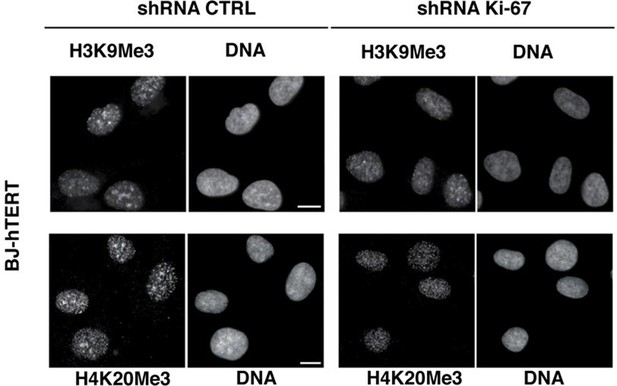
Heterochromatic histone mark localisation requires Ki-67.
Immunofluorescence analysis of H3K9me3 and H4K20me3 localisation in stable control and Ki-67 knockdown hTERT immortalised human fibroblasts (HDF BJ hTERT) cells. Scale bars, 10 µm.
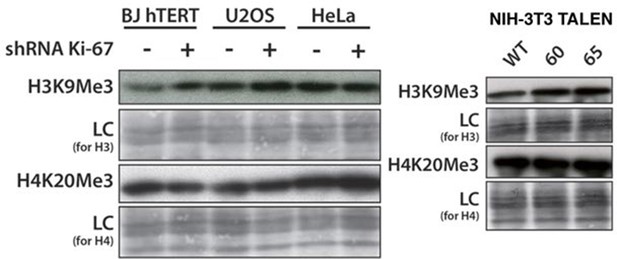
Overall heterochromatic histone mark levels do not change upon Ki-67 knockdown.
Left: western blot of total H3K9me3 and H4K20me3 level in control and stable Ki-67 knockdown BJ-hTERT, U2OS and HeLa cells. LC, loading control. Right: western blot of total H3K9me3 and H4K20me3 level in NIH-3T3 WT clone W4 and Ki-67-negative TALEN clones 60, 65. LC, loading control
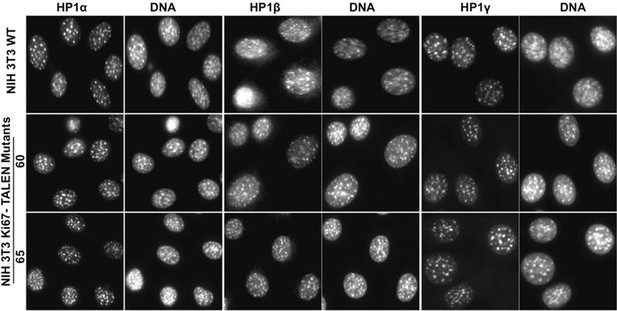
HP1 localises normally to chromocentres in Ki-67 mutant cells.
Immunofluorescence analysis of localisation of HP1α, HP1β and HP1γ in WT and Ki-67-negative NIH-3T3 mutant clones 60 and 65, as compared with DAPI staining.
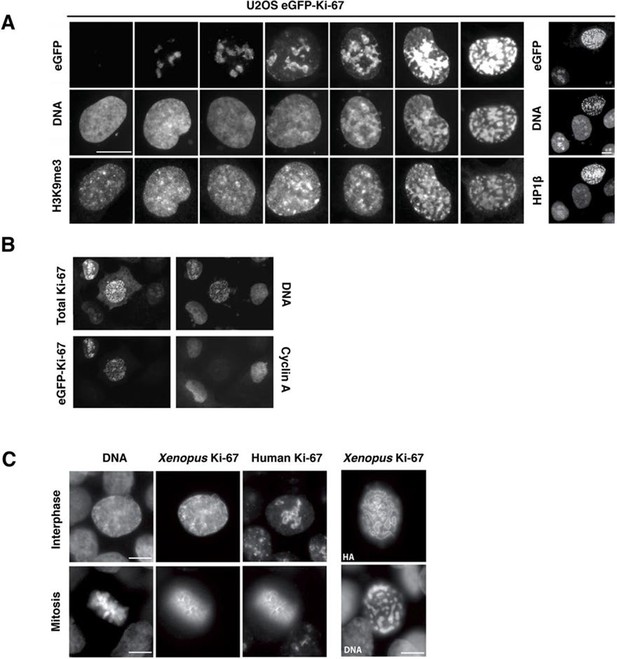
Overexpression of Ki-67 induces ectopic heterochromatin.
(A) Overexpression of full length Ki-67 N-terminal fusion with eGFP in U2OS cells induces ectopic heterochromatin, as visualised by DAPI staining (middle) and immunofluorescence of H3K9Me3 (left) or HP1β (right). Eight representative cells that have different levels of Ki-67 expression, as determined by eGFP fluorescence intensity, are shown. Bar, 10 µm. (B) U2OS cells expressing high levels of exogenous eGFP-Ki-67 and showing ectopic chromatin condensation are negative for cyclin A staining by immunofluorescence. (C) Left: Immunofluorescence analysis of the localisation of endogenous human and ectopically expressed Xenopus Ki-67 in U2OS cells, showing colocalisation in metaphase at the perichromosomal region. Right: DNA condensation caused by high overexpression of Xenopus Ki-67 in U2OS cells. Bars, 10 µm.
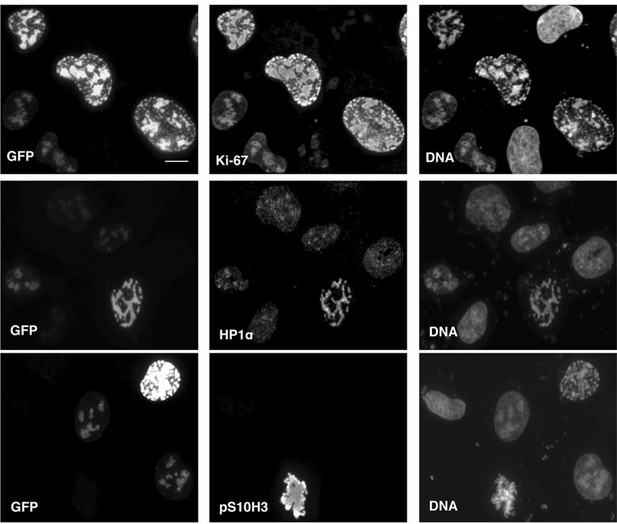
Overexpression of Ki-67 induces ectopic heterochromatin.
Overexpression of full length Ki-67 N-terminal fusion with eGFP (GFP, left panels) in U2OS cells induces ectopic heterochromatin, as visualised by DAPI staining (DNA, right panels) or immunofluorescence of HP1α (centre, middle), whereas cells with lower Ki-67 expression levels have normal chromatin. Immunofluorescence of Ki-67 (top, middle) shows colocalisation of fusion protein with overall Ki-67 pattern. Immunofluorescence of phospho-histone H3S10 shows expected staining of mitotic metaphase (bottom, middle panel) but no staining in a cell with ectopic heterochromatin (top right cell, same panel) due to Ki-67 overexpression (GFP, bottom left panel). Bar, 10 µm
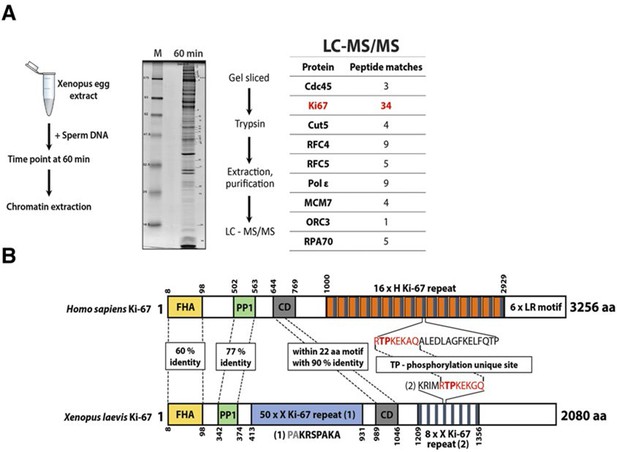
A Xenopus Ki-67 homologue (A) Chromatin proteomics in replicating Xenopus egg extracts.
Scheme of the experiment and electrophoresis separation of sample processed and analysed by mass spectrometry. Table shows peptide matches. M, molecular weight marker. (B) Schematic comparison of Xenopus Ki-67 homologue with human Ki-67 (long form). Domains are indicated by boxes (FHA, forkhead-associated domain; PP1, PP1-binding domain; CD, conserved domain). Highly conserved regions are indicated by dotted line with percentage of identical amino acids. TP, threonine-proline phosphorylation site.





