Lys29-linkage of ASK1 by Skp1−Cullin 1−Fbxo21 ubiquitin ligase complex is required for antiviral innate response
Figures
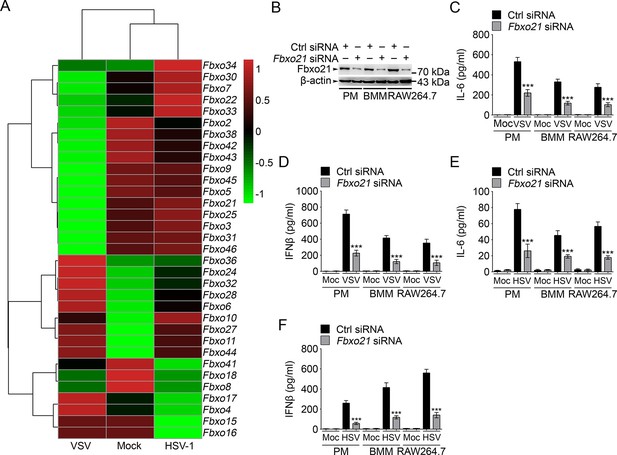
Knockdown of Fbxo21 inhibits innate antiviral response of macrophages.
(A) Heat map representation of F-box genes in RAW264.7 cells infected with VSV (MOI = 1) or HSV-1 (MOI = 5). (B) Efficiency of Fbxo21 knockdown in macrophages, as examined by immunoblot 48 hr after transient transfection of indicated control (Ctrl) or Fbxo21 siRNAs. One representative experiment of three was shown. PM, peritoneal macrophages; BMM, bone marrow-derived macrophages. (C–F) Cells (1 x 105 cells per 24-well) in (B) were infected with or without (Moc) VSV (C, D; MOI = 1) or HSV-1 (E, F; MOI = 5) as indicated for 8 hr. IL-6 (C, E) and IFNβ (D, F) concentrations in supernatant were determined by ELISA. Error bars indicated for mean ± SD of triplicate samples. Data were analyzed by one-way ANOVA followed by Bonferroni multiple comparison using PRISM software (***p < 0.001; versus control siRNA group infected with VSV or HSV-1).
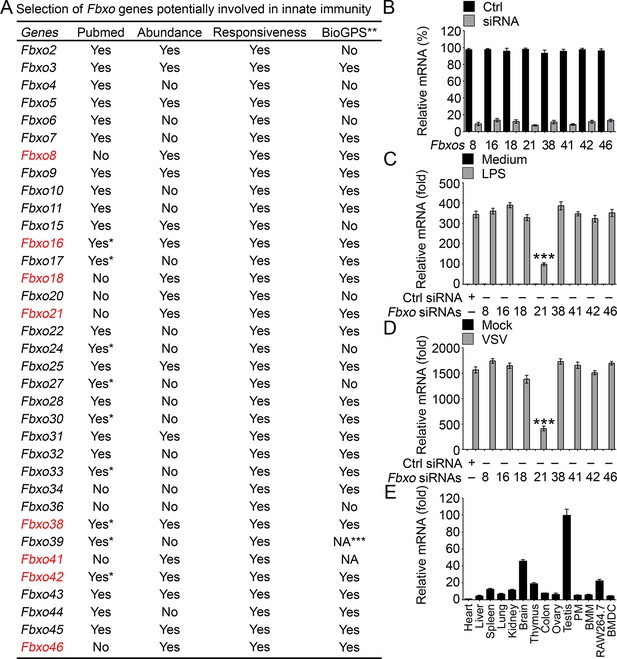
Identification of Fbxo21 as an important regulator of innate response.
(A) Selection of potential Fbxo molecules in innate immunity. '*' indicates that there are reports for indicated Fbxo proteins but no reports about their roles in innate immunity. '**' indicates for data of monocytes or RAW264.7 cells treated with LPS. '***' indicates that data are not available (NA). (B) Efficiency of indicated siRNAs was examined by Q-PCR. Error bars indicate for mean ± SD of triplicate samples. (C, D) Peritoneal macrophages were transiently transfected with control (Ctrl) siRNAs or indicated siRNAs for 48h, and then challenged with 100 ng/ml LPS (C, 6h) or VSV (MOI = 1, D; 8h). The mRNA levels of Ifnb were measured by Q-PCR. Error bars indicate for mean ± SD of triplicate samples. Data were analyzed by one-way ANOVA followed by Bonferroni multiple comparison using PRISM software (***, p < 0.001; versus control siRNA group treated with LPS or VSV). (E) Expression profiles of Fbxo21 mRNA in tissues and cells were examined by Q-PCR. Error bars indicate for mean ± SD of triplicate samples. PM, peritoneal macrophages; BMM, bone marrow-derived macrophages; BMDC, bone marrow-derived dendritic cells. β-actin was used as internal control in the Q-PCR assays.
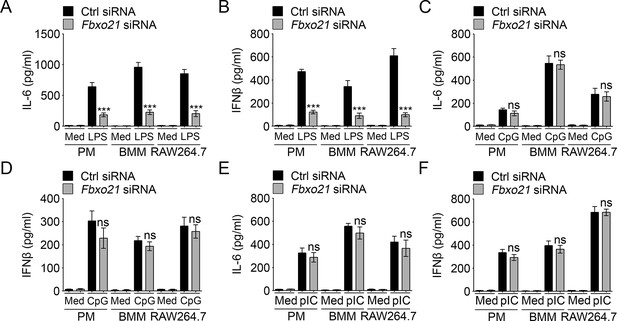
Knockdown of Fbxo21 impairs innate TLR4 response.
48 hr after transient transfection of indicated control (Ctrl) or Fbxo21 siRNAs, cells (1 × 105 cells per 24-well) were treated with or without 100 ng/ml LPS (A, B), 1 μM CpG-ODN (C, D) or 10 μg/ml poly (I:C) (E, F) as indicated for 6 hr. IL-6 (A, C, E) and IFNβ (B, D, F) concentrations in supernatant were determined by ELISA. Error bars indicated for mean ± SD of triplicate samples. Data were analyzed by one-way ANOVA followed by Bonferroni multiple comparison using PRISM software (ns, not significant; ***p < 0.001; versus control siRNA group treated with TLR agonists). Med, medium; CpG, CpG-ODN; pIC, poly (I:C).
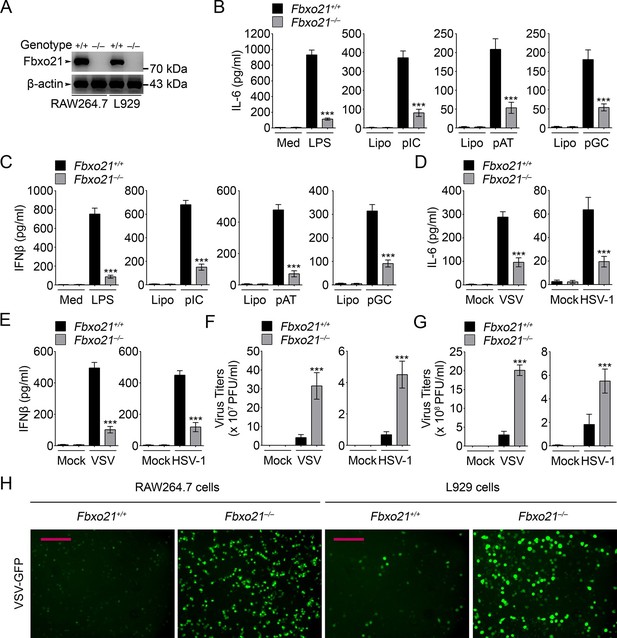
Deficiency of Fbxo21 impairs antiviral innate response.
(A) Confirmation of Fbxo21 depletion. Fbxo21 expression in single-clone-derived RAW264.7 cells and L929 cells established by CRISPR-Cas9 editing was examined by immunoblot. One representative experiment of three was shown. (B, C) Fbxo21+/+ and Fbxo21-/- RAW264.7 cells (1 × 105 cells per 24-well) were treated with or without 100 ng/ml LPS (for 6 hr), or transfected with 2.5 μg/ml of liposome-packaged poly (I:C), poly (dA:dT) or poly (dG:dC) (for 8 hr). IL-6 (B) and IFNβ (C) concentrations in supernatant were determined by ELISA. Med, medium; CpG, CpG-ODN; pIC, poly (I:C). (D–G) Fbxo21+/+ and Fbxo21-/- RAW264.7 cells (D–F) or L929 cells (G) were infected with or without (Mock) VSV (MOI = 1) or HSV-1 (MOI = 5) as indicated for 8 hr (D, E) or 12 hr (F, G). IL-6 (D) and IFNβ (E) concentrations in supernatant were determined by ELISA. The titers of viruses in cells were determined by plaque formation assay (F, G). In (B–G), error bars indicated for mean ± SD of triplicate samples. Data were analyzed by one-way ANOVA followed by Bonferroni multiple comparison using PRISM software (ns, not significant; ***p < 0.001; versus corresponding Fbxo21+/+ group). (H) Fbxo21+/+ and Fbxo21-/- RAW264.7 cells were infected with or without (Mock) VSV-GFP virus (MOI = 1) for 8 hr. Then cells were viewed under immunofluorescence microscope. One representative experiment of three was shown. Scale bars: 200 μm.
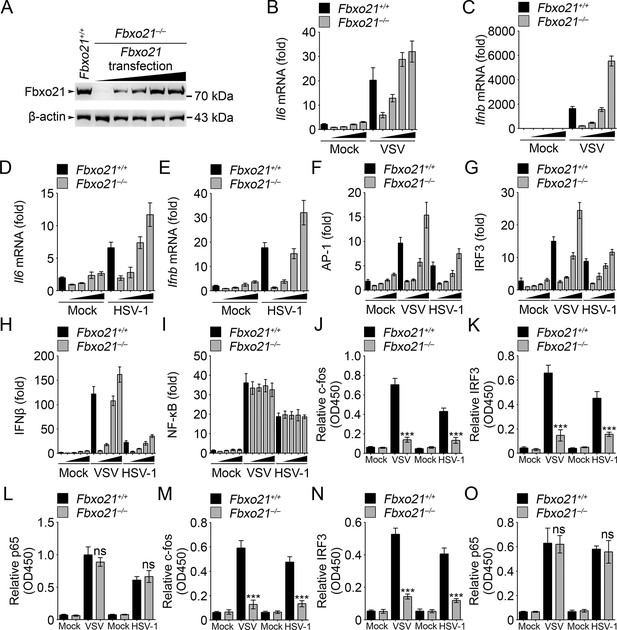
Fbxo21 overexpression promotes innate antiviral response, and activates AP-1 and IRF3-IFNβ signaling pathway.
(A) Fbxo21-/- RAW264.7 cells were transfected with Fbxo21 vectors (0, 0.1, 0.2, 0.5 and 1 μg respectively) for 48 hr. Fbxo21 expression was evaluated by immunoblot. (B–E) Cells in (A, transfected with 0, 0.1, 0.2 and 0.5 μg Fbxo21 vectors respectively) were infected with VSV (MOI = 1) or HSV-1 (MOI = 5) for 8 hr. Then indicated cytokines were examined by Q-PCR. β-actin was used as internal control in the Q-PCR assays. (F–I) Cells in (A, transfected with 0, 0.1, 0.2 and 0.5 μg Fbxo21 vectors respectively) were also transfected with indicated reporters and infected with VSV or HSV-1 for 4 hr. Then reporter transactivation was measured by dual-luciferase activity assays. (J–O) Fbxo21+/+ and Fbxo21-/- RAW264.7 cells (J–L) or L929 cells (M–O) were infected with VSV (MOI = 1) or HSV-1 (MOI = 5) for 4 hr. Then nuclear extracts were examined for indicated transcription factors bound to specific DNA sequence by ELISA. Error bars indicated for mean ± SD of triplicate samples. Data were analyzed by one-way ANOVA followed by Bonferroni multiple comparison using PRISM software (ns, not significant; ***p<0.001; versus corresponding Fbxo21+/+ group).
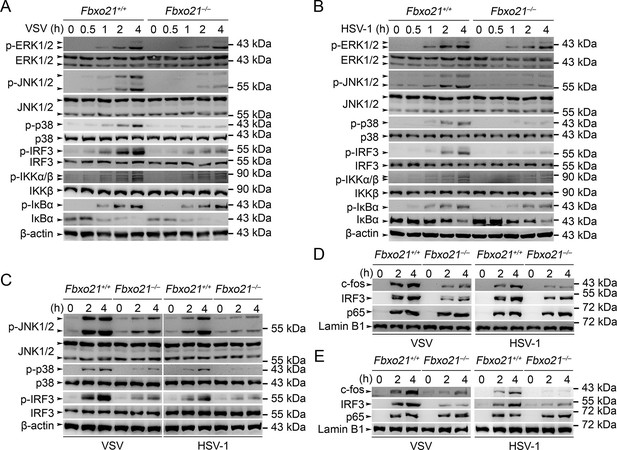
Deficiency of Fbxo21 inhibits virus-induced activation of JNK/p38-AP-1 and IRF3.
(A–C) Fbxo21+/+ and Fbxo21-/- RAW264.7 cells (A, B) or L929 cells (C) were infected with VSV (MOI = 1) or HSV-1 (MOI = 5) as indicated. The phosphorylation of indicated molecules was examined by immunblot. (D, E) Fbxo21+/+ and Fbxo21-/- RAW264.7 cells (D) or L929 cells (E) were treated as in (A–C), and the nuclear proteins were extracted and examined for indicated transcription factors by immunoblot. One representative experiment of three was shown.
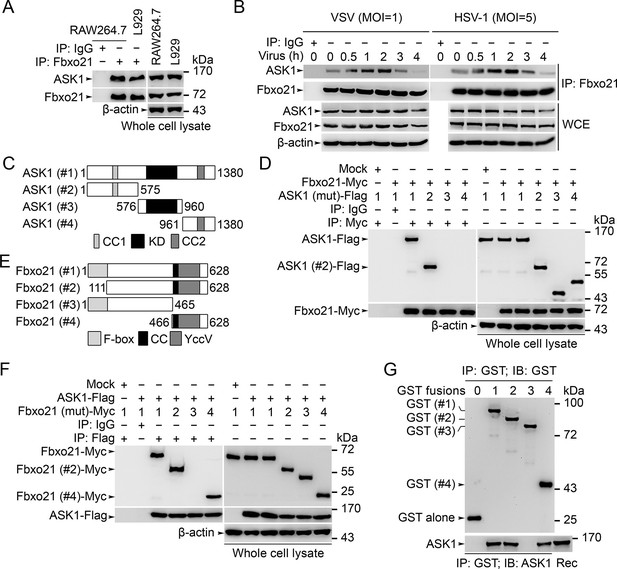
Fbxo21 interacts with ASK1.
(A, B) Wild type RAW264.7 cells or L929 cells infected with (B) or without (A) VSV and HSV-1 were prepared for whole cell extracts (WCE), which were immunoprecipitated (IP) with anti-Fbxo21 or IgG as indicated. The associated ASK1 was examined by immunoblot. (C) Schema of ASK1 mutants. CC1, coiled-coil domain 1; KD, kinase domain; CC2, coiled-coil domain 2. (D) HEK293 cells were cotransfected with Fbxo21-Myc and indicated Flag-tagged ASK1 mutants (1–4 mutants as illustrated in (C)) for 48 hr. Whole cell lysates immunoprecipitated (IP) with anti-Myc agaroses were examined for Flag-tagged ASK1 (mutant) by immunoblot. (E) Schema of Fbxo21 mutants. F-box, F-box domain; CC, coiled-coil domain; YccV, YccV domain. (F) HEK293 cells were cotransfected with ASK1-Flag and indicated Myc-tagged Fbxo21 mutants (1–4 mutants as illustrated in (E)) for 48 hr. Whole cell lysates immunoprecipitated (IP) with anti-Flag agaroses were examined for Myc-tagged Fbxo21 (mutant) by immunoblot. (G) GST pull-down assays of Fbxo21 interaction with recombinant (Rec) ASK1. '0' indicates for GST alone, and 1–4 indicate for GST-fused Fbxo21 (mutant) as in (E). One representative experiment of three was shown.
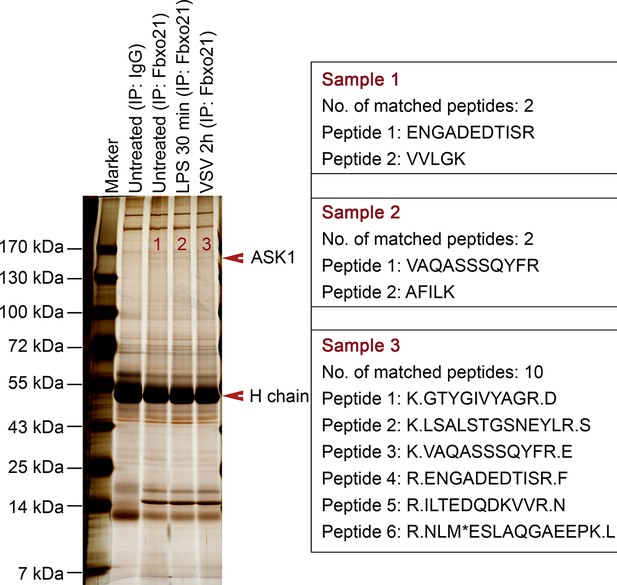
MS assays of Fbxo21-associated proteins.
Wild type RAW264.7 cells were treated with or without 100 ng/ml LPS or VSV (MOI = 1) as indicated. Whole cells lysates were immunoprecipitated (IP) with anti-Fbxo21 antibody plus protein A/G beads. The immune complexes were separated in a SDS-PAGE gel, and stained with silver buffer. The differential bands were subjected to MS assays. The identified unique peptides of ASK1 were shown at the right side.
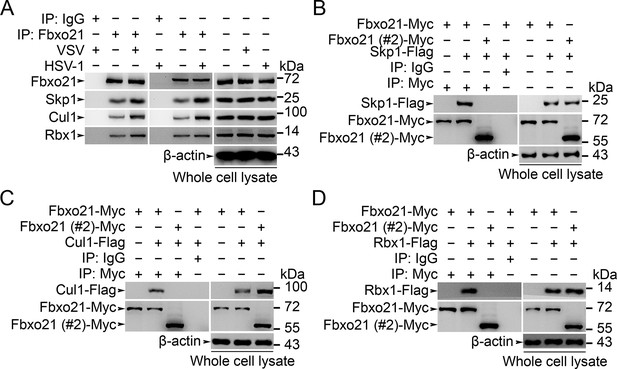
Fbxo21 is a component of SCF complex.
(A) Wild type RAW264.7 cells were treated with or without VSV (MOI = 1) or HSV-1 (MOI = 5) as indicated for 2 hr. Whole cell lysates immunoprecipitated (IP) with anti-Fbxo21 antibody plus protein A/G agaroses were examined for indicated SCF components by immunoblot (IB). (B–D) HEK293 cells were cotransfected with Myc-tagged Fbxo21 or Fbxo21 without F-box domain (#2) and indicated Flag-tagged SCF components for 48 hr. Whole cell lysates immunoprecipitated (IP) with anti-Myc agaroses were examined for indicated Flag-tagged proteins by immunoblot (IB). One representative experiment of three was shown.
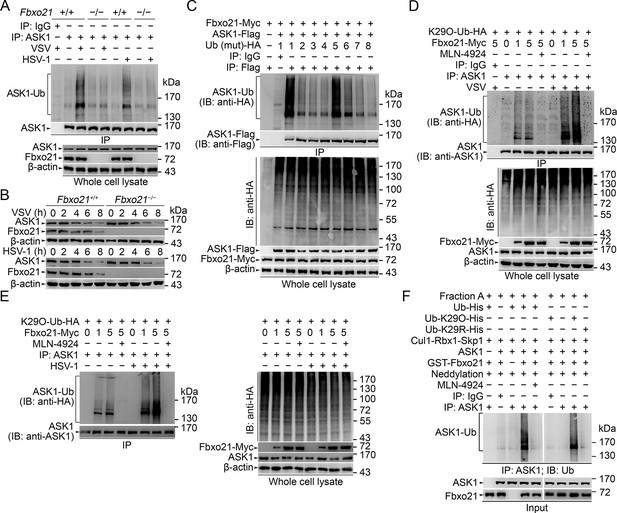
SCFFbxo21 mediates virus-induced Lys29-linkage of ASK1.
(A) Fbxo21+/+ and Fbxo21-/- RAW264.7 cells were infected with VSV (MOI=1) or HSV-1 (MOI = 5) for 2 hr. Then polyubiquitination of ASK1 was examined by immunoblot (IB) after immunoprecipitations (IP). (B) Fbxo21+/+ and Fbxo21-/- RAW264.7 cells were infected with VSV (MOI = 1) or HSV-1 (MOI = 5) as indicated. ASK1 and Fbxo21 levels in whole cell lysates were examined by immunoblot. (C) Myc-tagged Fbxo21 and Flag-tagged ASK1 were cotransfected with HA-tagged Ub (mutant) into HEK293 cells for 48 hr, and infected with VSV (MOI = 1) for 2 hr. Then polyubiquitinated ASK1 was examined by immunoblot (IB) against HA after immuneprecipitations (IP) with anti-Flag agaroses. 1, wild type Ub; 2, Ub with only the Lys6 residue unchanged (K6O-Ub); 3, K11O-Ub; 4, K27O-Ub; 5, K29O-Ub; 6, K33O-Ub; 7, K48O-Ub; 8, K63O-Ub. (D, E) Fbxo21-/- RAW264.7 cells were transfected with indicated amounts (0, 1 or 5 μg) Fbxo21-Myc and equal amounts of K29O-Ub-HA vectors for 48 hr, and then infected with VSV (MOI = 1) or HSV-1 (MOI = 5) for 2 hr in the presence or absence of MLN-4924 (10 nM). Then polyubiquitinated ASK1 was examined by immunoblot (IB) against HA after immuneprecipitations (IP). (F) After incubation for 30 min, the in vitro polyubiquitination system was boiled for 5 min, and then polyubiquitinated ASK1 was examined by immunoblot (IB) against Ub after immuneprecipitations (IP). One representative experiment of three was shown.
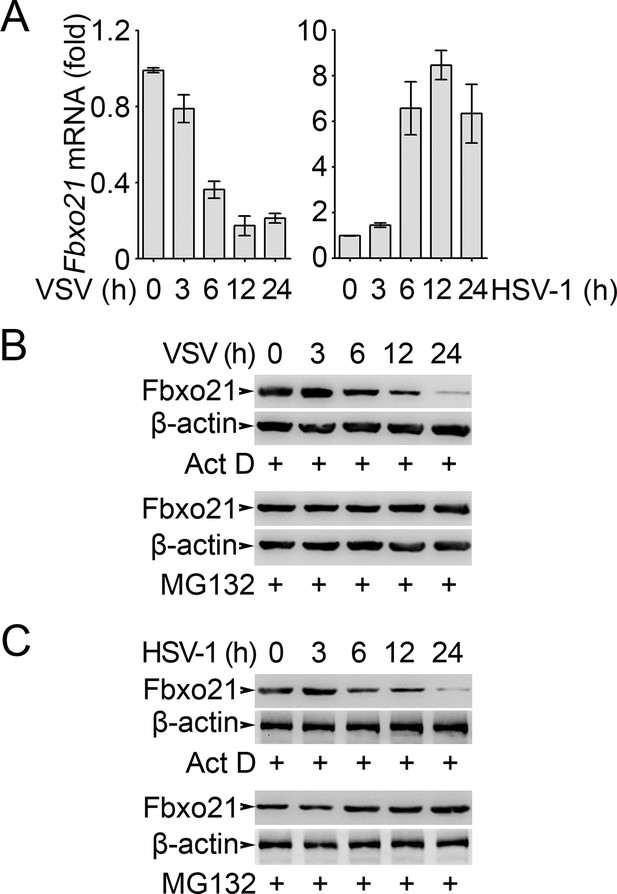
Analysis of Fbxo21 expression and degradation.
RAW264.7 cells were infected with VSV (MOI = 1) or HSV-1 (MOI = 5) as indicated in the presence (B, C) or absence (A) of actinomycin D (Act D, 10 μM) or MG132 (10 μM). Then the mRNA level of Fbxo21 (A, error bars indicate for mean ± SD of triplicate samples) and the protein levels of Fbxo21 (B, C) were examined by Q-PCR and Western blot, respectively. β-actin was used as internal control in the Q-PCR assays. One representative experiment of three was shown.
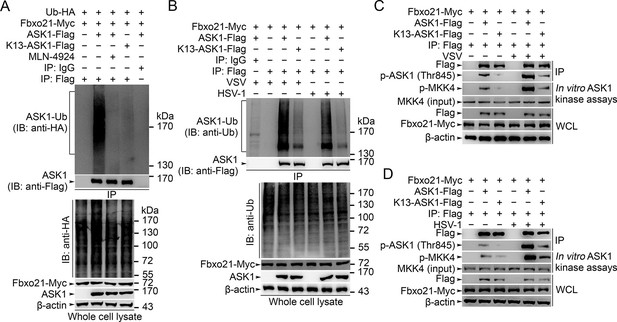
Lys29-linkage of ASK1 is crucial for Fbxo21-mediated ASK1 activation.
(A) HEK293 cells transiently transfected with indicated vectors for 48 hr and infected with VSV (MOI = 1) for 2 hr in the presence or absence of MLN-4924 (10 nM). Then ASK1 (mutant) was immunoprecipitated (IP) with anti-Flag agaroses, and polyubiquitination was evaluated by immunblot (IB) against HA. (B) Fbxo21–/– RAW264.7 cells were transfected with indicated vectors for 48 hr and infected with VSV (MOI = 1) or HSV-1 (MOI = 5) for 2 hr. Then ASK1 (mutant) was immunoprecipitated (IP) with anti-Flag agaroses, and polyubiquitination was evaluated by immunblot (IB) against Ub. (C, D) Fbxo21-/–- RAW264.7 cells were transfected with indicated vectors for 48 hr and infected with VSV (MOI = 1, C) or HSV-1 (MOI = 5, D) for 2 hr. Then ASK1 (mutant) was immunoprecipitated (IP) with anti-Flag agaroses, and the phosphorylated ASK1 was examined by immunoblot. Otherwise, the ASK1 (mutant) after IP was incubated with recombinant MKK4, and then immunoblotted for phosphorylated MKK4. One representative experiment of three was shown.
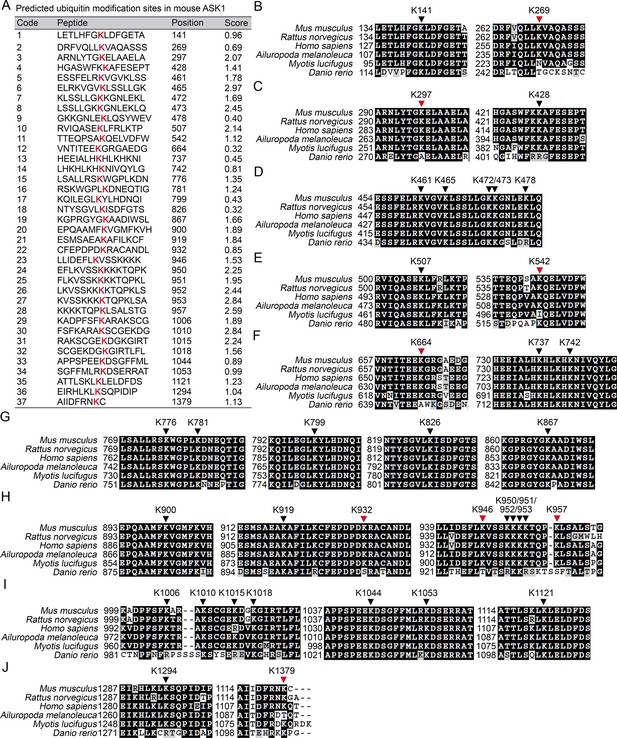
Prediction of the Lys residue in ASK1 responsible for linkage with Ub chain.
(A) The predicted Ub modification sites in ASK1 by using the Bayesian discriminant method-prediction of ubiquitination sites algorithm. (B–J) Alignment of ASK1 protein sequences derived from multiple species. Black arrow heads indicate for conserved Lys residues, and the red arrow heads for not completely conserved Lys residues.
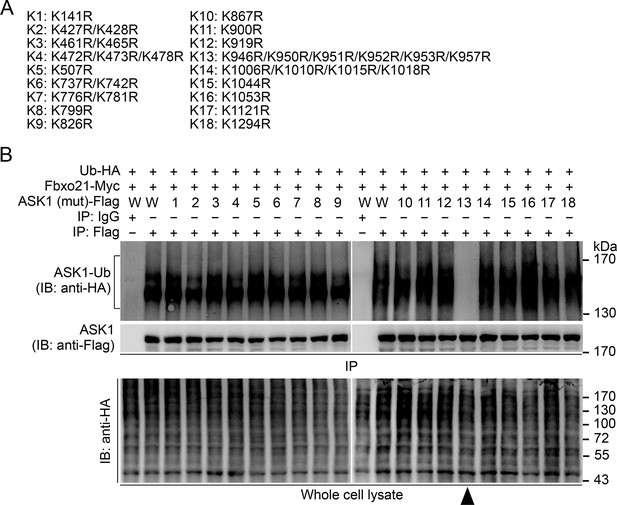
Confirmation of the Lys residue in ASK1 responsible for linkage with Ub chain.
(A) Summary of the mutated ASK1 with indicated Lys residue(s) mutated into Arg. (B) HEK293 cells were transiently transfected with indicated vectors for 48 hr and infected with VSV (MOI = 1) for 2 hr. Then Polyubiquitination of ASK1 (mutant) in immunoprecipitations (IPs) was evaluated by immunoblot (IB). Arrow head indicates for the K13-ASK1. 1 to 18 mutants of ASK1 were corresponding to those in (A, K1 to K18). W, wild type ASK1. Prediction of the Lys residue in ASK1 responsible for linkage with Ub chain. One representative experiment of three was shown.
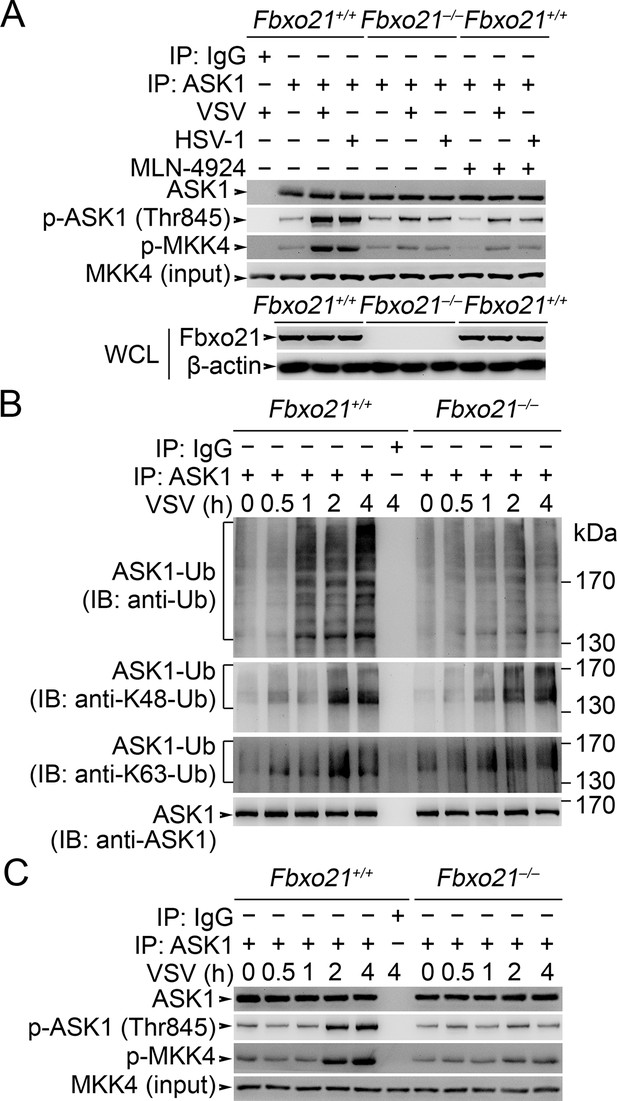
Fbxo21 deficiency impairs poly-Ub modification of ASK1 and then ASK1 activation.
(A) Fbxo21+/+ or Fbxo21–-/- RAW264.7 cells were infected with VSV (MOI = 1) or HSV-1 (MOI = 5) for 2 hr in the presence or absence of MLN-4924 (10 nM). Then ASK1 was immunoprecipitated (IP) with anti-ASK1 antibody plus protein A/G agaroses, and the phosphorylated ASK1 was examined by immunoblot. Otherwise, the ASK1 after IP was incubated with recombinant MKK4, and then immunoblotted for phosphorylated MKK4. (B, C) Fbxo21+/+ or Fbxo21-/-RAW264.7 cells were infected with VSV (MOI = 1) as indicated. Then the polyubiquitination of ASK1 (B) and activation of ASK1 (C, examined as in A) were analyzed after immunoprecipitations (IP). One representative experiment of three was shown.
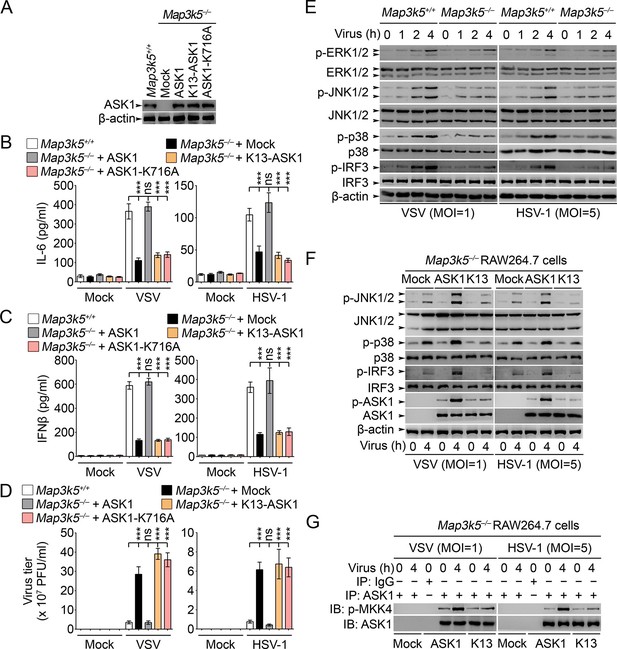
Lys29-linkage of ASK1 is crucial for innate antiviral response.
(A) Map3k5+/+ RAW264.7 cells and Map3k5–/– RAW264.7 cells transfected with mock vector or indicated ASK1 vectors for 48 hr were examined for ASK1 expression by Western blot. (B–D) Map3k5+/+ and Map3k5–/– RAW264.7 cells (as in A) were infected with or without (Mock) VSV (MOI = 1) or HSV-1 (MOI = 5) as indicated for 8 hr (B, C) or 12 hr (D). IL-6 (B) and IFNβ (C) concentrations in supernatant were determined by ELISA. The titers of viruses in cells were determined by plaque formation assay (D). Error bars indicated for mean ± SD of triplicate samples. Data were analyzed by one-way ANOVA followed by Bonferroni multiple comparison using PRISM software (ns, not significant; ***p < 0.001). (EG) Cells in (A) were infected with or without (Mock) VSV (MOI = 1) or HSV-1 (MOI = 5) as indicated. Then whole cell lysate (E, F) or immunoprecipitates (G) were examined by Western blot. In (G), the immunoprecipitated ASK1 was examined for the kinase activity in the presence of 10 ng recombinant MKK4. One representative experiment of three was shown.
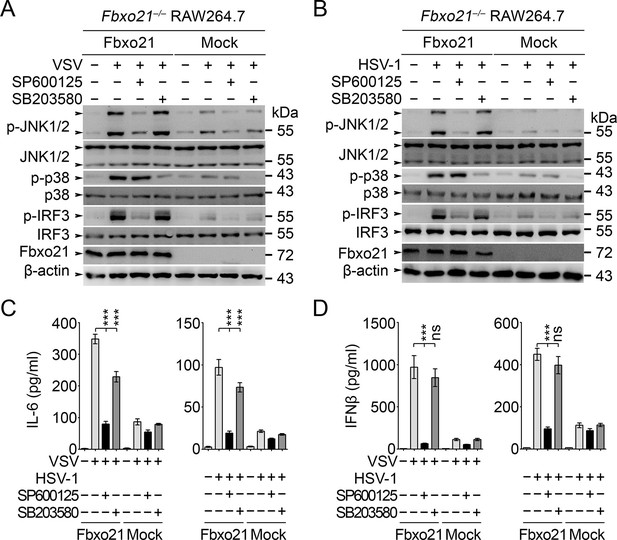
JNK1/2 activation is crucial for Fbxo21-mediated innate response.
Fbxo21–/– RAW264.7 cells transfected with mock or Fbxo21 vectors were infected with VSV (MOI = 1) or HSV-1 (MOI = 5) for 4 hr (A, B) or 8 hr (C, D) in the presence or absence of SP600125 (20 μM) or SB203580 (20 μM). The indicated proteins were examined by immunoblot (A, B), and the cytokine concentrations in the supernatant were measured by ELISA (C, D). One representative experiment of three was shown (A, B). Error bars indicated for mean ± SD of triplicate samples (C, D). Data were analyzed by one-way ANOVA followed by Bonferroni multiple comparison using PRISM software (ns, not significant; ***p < 0.001).
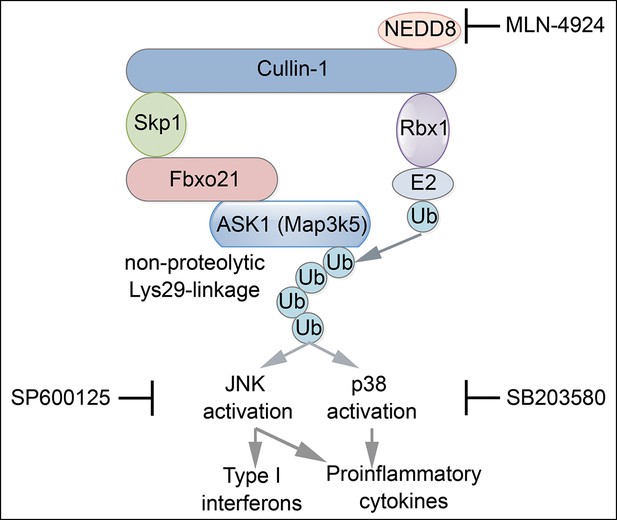
Proposed working model for Fbxo21-mediated innate antiviral response.
Upon infection with RNA or DNA viruses, host cells can recognize nucleic acids of viruses and initiate MAVS- or STING-dependent signaling pathway to induce the production of inflammatory cytokines and type I interferons. When Fbxo21 is involved, it can mediate Lys29-linkage of ASK1 by joining with the SCF complex, and in turn, activate JNK1/2 (inhibited by SP600125) and p38 (inhibited by SB203580), leading to elevated activation of AP-1 and IRF3 for induction of the cytokines.





