Spatial control of translation repression and polarized growth by conserved NDR kinase Orb6 and RNA-binding protein Sts5
Figures
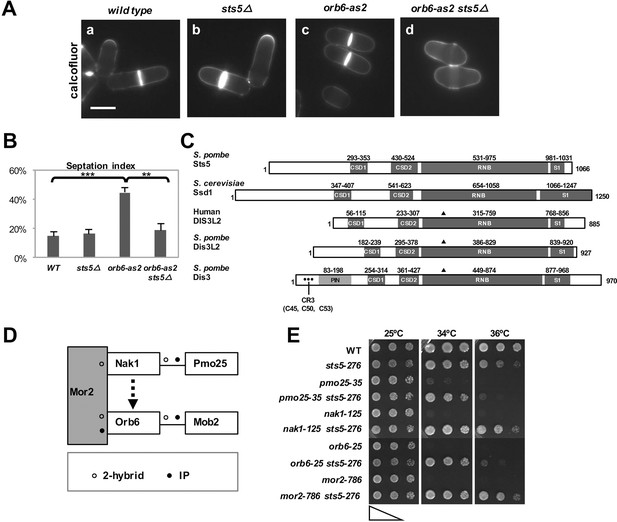
Loss of RNA-binding protein Sts5 suppresses the cell viability defects of orb6 mutants.
(A) Deletion of sts5 suppresses the cell separation phenotype of analog-sensitive orb6-as2 mutants. (a) wild-type, (b) sts5∆, (c) orb6-as2 and (d) orb6-as2 sts5∆ mutants treated with 50 μM 1-NA-PP1 inhibitor for 2 hr at 32°C. Bar = 5 μm. (B) Septation index quantification of cells in experiment shown in A based on 3 independent experiments (N>295 per strain). Orb6-as2 cells exhibit a significantly higher septation index as compared to control cells (P = 0.0004) and as compared to orb6-as2 sts5∆ double mutants (P = 0.0011). P values were determined using analysis of variance (ANOVA) with SPSS statistics package 22.0, followed by Tukey’s HSD test. Error bars indicate SD. (C) Sts5 protein sequence includes the RNB domain (a.a. 531–975), with homology to the catalytic domain of E. coli ribonuclease II, three conserved OB-fold domains that promote interaction with RNA, the CSD1 (a.a. 293–353), CSD2 (a.a. 430–524), and S1 (a.a. 981–1031) domains. Sts5 is related to the exoribonuclease Dis3L2 that is conserved from S. pombe to humans. ▲ indicates 3 catalytic residues in RNB domain. • indicates CR3 motif residues for exosome targeting. (D) Interactions between MOR network proteins. Mor2 serves as a scaffold that enables activation of Mob2-bound Orb6 by the Nak1-Pmo25 complex (o indicates 2-hybrid interaction; • indicates IP interaction). (E) sts5-276 mutation suppresses the temperature-sensitive growth of MOR mutants. The indicated cells were spotted on YPD solid medium (approximately 5 × 104 cells in the left spots for each plate and then diluted 4-fold in each subsequent spot) and incubated at 25°C, 34°C, and 36°C for 3 days.
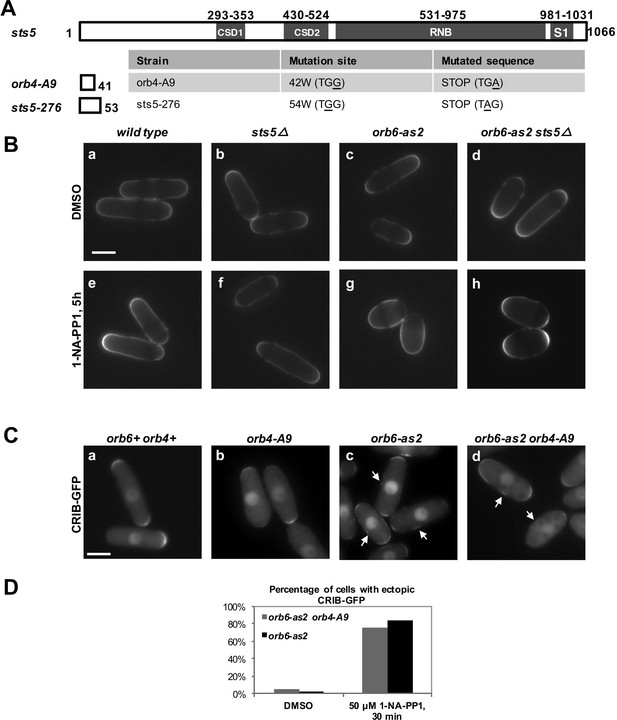
Loss of RNA-binding protein Sts5 does not suppresses the polarity defects observed upon Orb6-as2 kinase inhibition.
(A) Description of orb4-A9 and sts5-276 early stop codon mutations in the sts5 gene. No significant differences were observed in the phenotype of these mutations as compared to the sts5∆ deletion. (B) Loss of sts5 (sts5∆) does not suppress the cell polarity phenotype of orb6-as2 cells following Orb6 kinase inhibition for 5 hr. Bar = 5 μm. (C) Loss of sts5 (orb4-A9 allele) does not suppress CRIB-GFP mislocalization in orb6-as2 mutants. Cells were grown at 32°C and treated with 1-NA-PP1 inhibitor for 30 mins. Bar = 10 μm. Note that only the subset of cells that do not contain a septum (non-septating cells) were analyzed. (D) Quantification of CRIB-3xGFP localization as shown in (C) (N≥70 cells per condition).
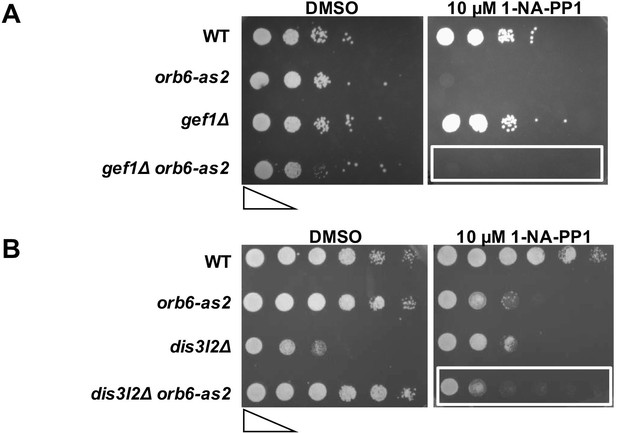
Deletion of gef1 or dis3L2 does not suppress the growth defect observed upon Orb6-as2 kinase inhibition.
(A) gef1Δ cells do not suppress the growth defect observed upon Orb6-as2 kinase inhibition. Growth properties of the gef1Δ orb6-as2 double mutant compared with wild-type, orb6-as2, and gef1Δ cells were assayed by spotting the indicated cells on minimal solid medium in the presence of DMSO or 10 μM 1-NA-PP1 inhibitor and incubated the plates at 32°C for 3 days (approximately 5 × 105 cells in the left spots for each plate and then diluted 10-fold in each subsequent spot). (B) Loss of the sts5 homologue dis3L2 does not suppress the growth defect observed upon Orb6-as2 kinase inhibition. Growth properties of the dis3L2Δ orb6-as2 double mutant compared with wild-type, orb6-as2, and dis3L2Δ cells were assayed by spotting the indicated cells on minimal solid agar as described in A.
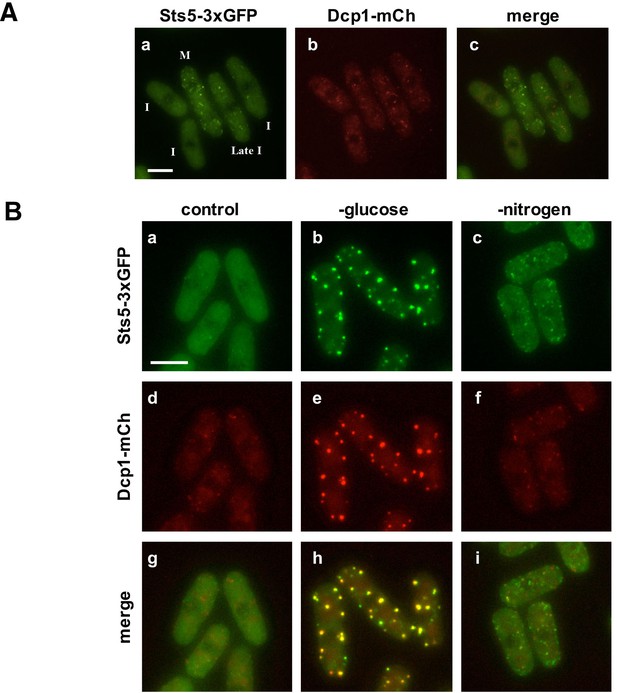
Sts5 proteins assemble into puncta during mitosis and during nutritional starvation.
(A) Sts5-3xGFP proteins coalesce into cytoplasmic particles in cells undergoing mitosis (M) but appear mostly diffuse in the cytoplasm of growing interphase (I) cells (a, c). (b) P-body formation, as visualized by P-body marker Dcp1-mCherry is not induced in mitotic cells. Bar = 5 μm. (B) Sts5-3xGFP proteins are recruited and colocalize with the P-body marker Dcp1-mCherry upon growth for 1 hr in minimal medium minus glucose (b, e, h). Sts5-3xGFP recruitment and colocalization with Dcp1-mCherry in P-bodies also occurs upon 1 hr of growth in minimal medium minus nitrogen (c, f, i). Sts5-3xGFP recruitment was observed as early as 15 min after transfer to glucose- or nitrogen-depleted medium. Images are deconvolved projections from 12 Z-stacks separated by a step size of 0.3 μm. Experiment was performed using prototrophic strain FV2267. Bar = 5 μm.
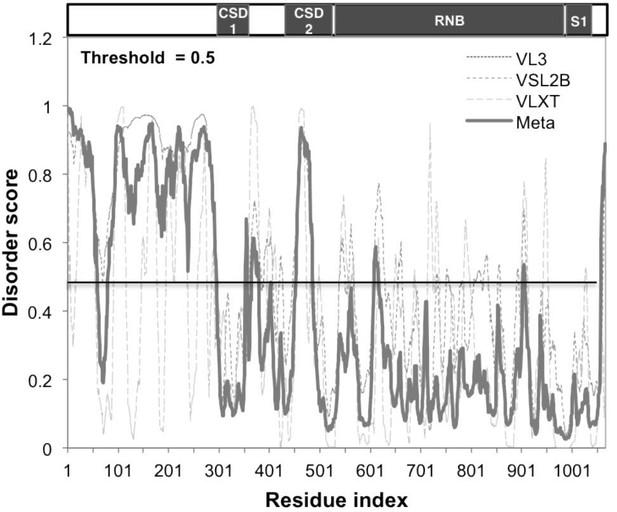
Sts5 protein contains an intrinsically disordered domain.
Sts5 protein contains an intrinsically disordered domain as predicted by DisProt software (using VL3, VSL2B, and VLXT algorithms) (DisProt - Database of Protein Disorder, RRID:SCR_007097).
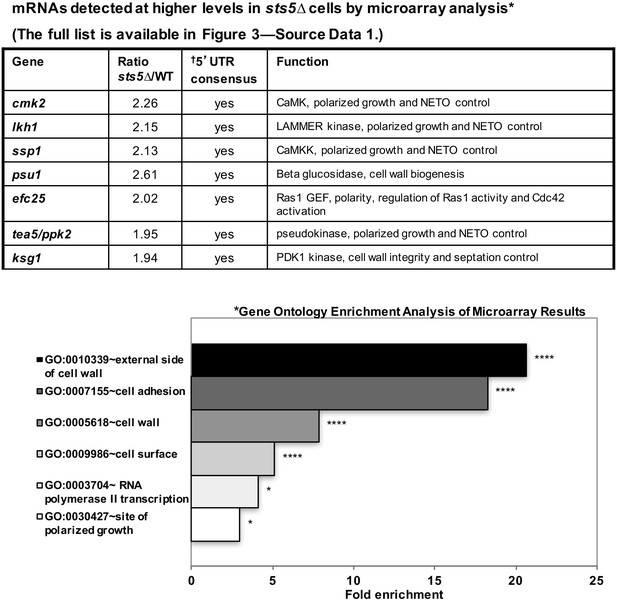
mRNAs detected at higher levels in sts5∆ cells by microarray analysis.
Total mRNA was extracted from sts5∆ and control cells for microarray analysis. A complete list of mRNAs increased in sts5∆ cells (≥1.9 fold) is shown in Figure 3—source data 1. Several of these mRNAs have established functions in bipolar growth activation and contain putative Sts5-binding sites in their 5’ UTRs (Hogan et al., 2008; Wanless et al., 2014). *Gene ontology enrichment analysis of terms that are significantly enriched among the set of mRNAs with sts5Δ/WT ratio ≥1.90 in the microarray results. Fold enrichment plotted per gene ontology category among all significant terms (P<0.05, modified Fisher Exact P-value with the Benjamini P-value correction) for Cellular Compartment (CC), Biological Process (BP) and Molecular Function (MF) Gene Ontology terms. †Sts5 binding site: HNNYAHTCHWW (where H = A,T,C / N = A,T,C,G / Y = C,U / W = T,A).
-
Figure 3—source data 1
Microarray analysis results.
A complete list of mRNAs detected at higher levels (≥1.9 fold) in sts5∆ cells by microarray analysis.
- https://doi.org/10.7554/eLife.14216.009
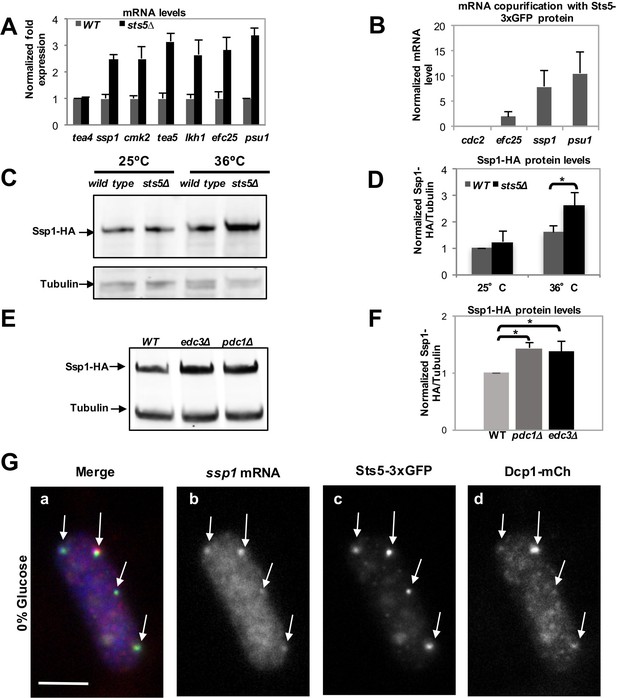
Loss of Sts5 leads to increased levels of mRNAs involved in growth control and bipolar growth activation.
(A) qPCR analysis confirmation that several of these transcripts are more abundant in the sts5∆ strain as compared to control cells based on 3 independent experiments. Tea4 is shown as an example of a transcript that is not altered. Housekeeping genes were nda3, act1, cdc2, and cdc22. Error bars indicate SD. (B) Interaction of ssp1, efc25, and psu1 mRNAs with Sts5-3xGFP as established by co-immunoprecipitation with Sts5-3xGFP followed by qPCR as described in the Materials and Methods. cdc2 is shown as an example of a transcript that does not interact with Sts5-3xGFP. Error bars indicate SD. Three independent experiments were performed. (C) Western blotting against Ssp1-HA performed as described in Materials and Methods in WT and sts5Δ cells cultured in YE medium at 25°C and 36°C. Tubulin levels were determined as a loading control. (D) Quantification of Ssp1-HA/Tubulin ratio normalized to WT levels was based on 3 independent experiments. Change in Ssp1-HA level is significantly greater in sts5∆ cells as compared to controls at 36°C (P = 0.034, Student’s t-test). Error bar=SD. (E) Western blotting against Ssp1-HA performed as described in Materials and Methods in WT and edc3Δ and pdc1Δ cells cultured in supplemented minimal medium at 30°C. Tubulin levels were determined as a loading control. (F) Quantification of Ssp1-HA/Tubulin ratio normalized to WT levels at 25°C based on 3 independent experiments. Change in Ssp1-HA level is significantly greater in edc3Δ (P = 0.018, Student’s t-test) and pdc1Δ (P = 0.0154, Student’s t-test) cells as compared to controls. Error bar = SD. (G) RNA FISH visualization of ssp1 mRNA in fixed cells cultured for 20 min in supplemented minimal medium containing 0% glucose. Hybridization used 20-mer DNA oligos (Stellaris) labeled with Quasar 705 fluorochromes. Bar = 5 μm.
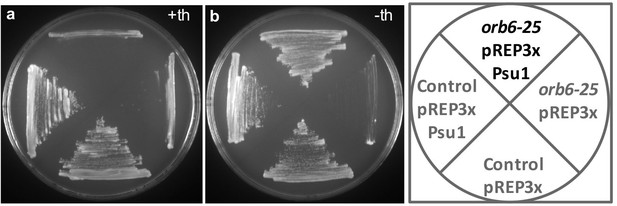
Overexpression of Psu1 suppresses the temperature-sensitive growth defect of orb6-25 mutant cells.
Psu1 was expressed in orb6-25 and control cells from the pRep3X plasmid under the control of the nmt1 promoter, at restrictive temperature (36°C), in the presence (a) or absence (b) of Thiamine.
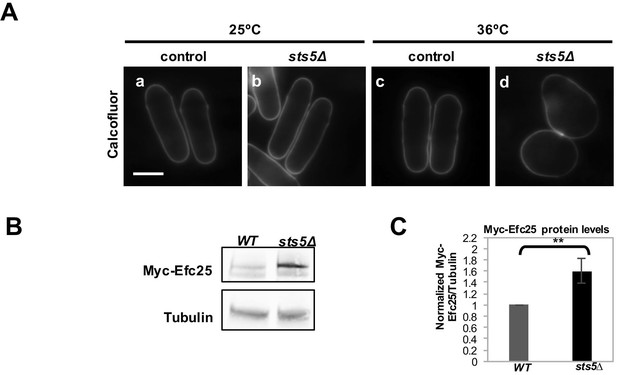
Deletion of sts5 alters cell shape and Myc-Efc25 protein levels.
(A) Calcofluor staining of WT (a and c) and sts5∆ (b and d) cultured in YE medium at 25°C and 36°C. Bar = 5 μm. (B) Western blotting against Myc-Efc25 performed as described in Materials and Methods in WT and sts5Δ cells cultured in minimal medium at 25°C. Tubulin levels were determined as a loading control. (C) Quantification of Myc-Efc25/Tubulin ratio normalized to WT levels at 25°C based on 3 independent experiments. Change in Myc-Efc25 level is significantly greater in sts5Δ cells as compared to controls (P = 0.0087, Student’s t-test).
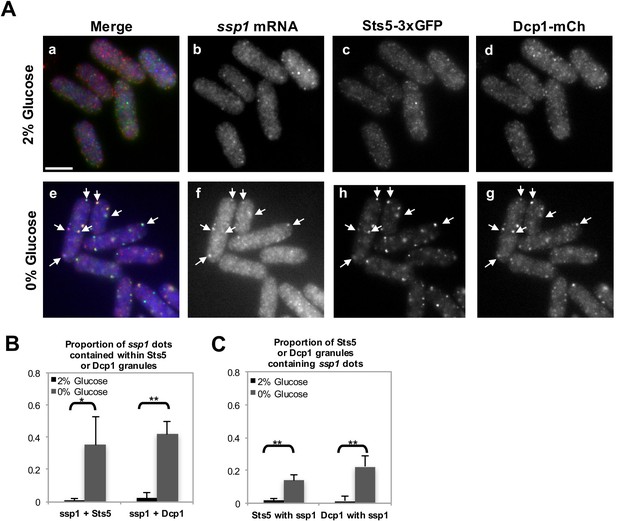
Extent of colocalization between ssp1 mRNA, Sts5-3xGFP, and Dcp1-mCherry in fixed cells cultured in the presence and absence of glucose.
(A) RNA FISH visualization of ssp1 mRNA in fixed cells cultured for 20 min in supplemented minimal medium containing 2% glucose or 0% glucose. Hybridization used 20-mer DNA oligos (Stellaris) labeled with Quasar 705 fluorochromes. Bar = 5 μm. (B) Object-based quantification of the proportion of ssp1 puncta contained within Sts5-3xGFP or Dcp1-mCherry puncta in the presence and absence of glucose. For ssp1 dots contained within Sts5 granules in 0% glucose vs 2% glucose, P = 0.0293 (Student’s t-test). For ssp1 dots contained within Dcp1 granules in 0% glucose vs 2% glucose, P = 0.0011 (Student’s t-test). Object-based quantification based on the distance between centers of mass performed using ImageJ plugin JACoP (Bolte and Cordelières, 2006). Error bars denote SD among the means of 3 independent experiments (N≥25 cells). For details, see Materials and methods. (C) Object-based quantification of the proportion of Sts5-3xGFP or Dcp1-mCherry puncta containing ssp1 puncta in the presence and absence of glucose. For Sts5 granules containing ssp1 dots in 0% glucose vs 2% glucose, P = 0.0042 (Student’s t-test). For Dcp1 granules containing ssp1 dots in 0% glucose vs 2% glucose, P = 0.0053 (Student’s t-test). Object-based quantification based on the distance between centers of mass performed using ImageJ plugin JACoP (Bolte and Cordelières, 2006). Error bars denote SD among the means of 3 independent experiments (N≥25 cells). For details, see Materials and Methods.
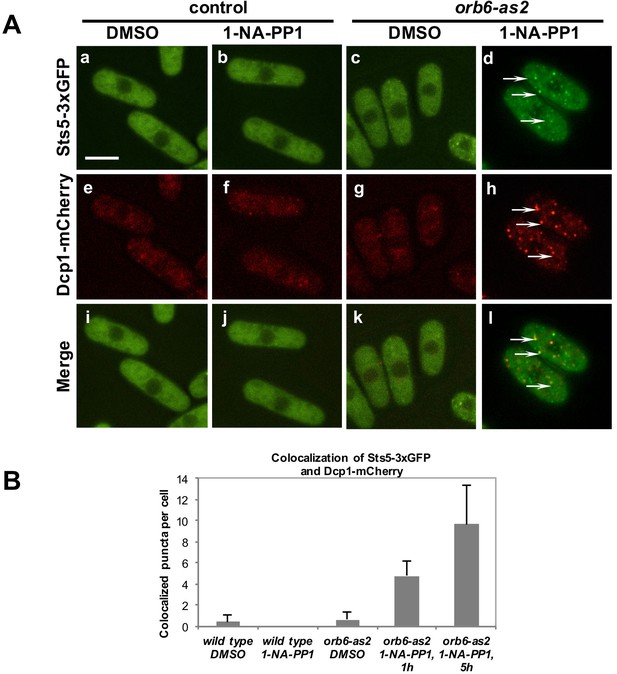
Orb6 kinase inhibits Sts5 recruitment and localization to P-bodies.
(A) Loss of Orb6-as2 kinase activity leads to Sts5-3xGFP recruitment into puncta that colocalize with the P-body marker Dcp1-mCherry. Cells were treated with inhibitor or DMSO for 1 hr (shown) and 5 hr. Bar = 5 μm. (B) Quantification of three sets of experiments as shown in A.
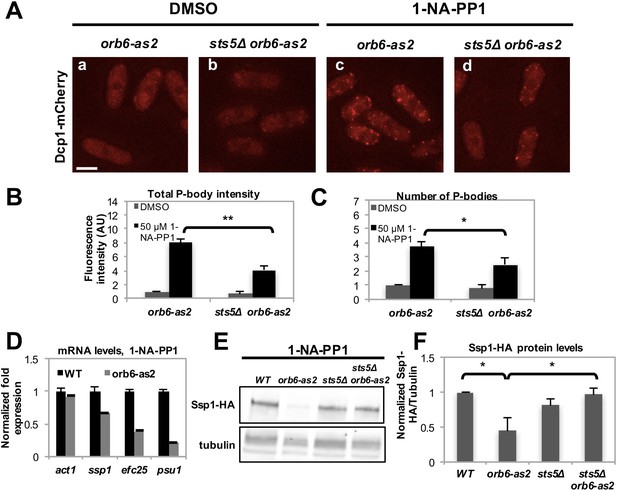
Orb6 kinase inhibits Sts5-dependent P-body formation and translational repression.
(A) Dcp1-mCherry localization in orb6-as2 (a, c) compared with sts5Δ orb6-as2 (b, d) cells grown in supplemented minimal medium in the presence of 50 μM 1-NA-PP1 (c and d) or DMSO (a and b) for 1 hr. Loss of Sts5 in the sts5∆ orb6-as2 strain decreases the number of P-bodies induced by Orb6 kinase inhibition. Images are deconvolved projections from 12 Z-stacks separated by a step size of 0.3 μm. Bar = 5 μm. (B) Quantification of the experiment shown in A based on 3 independent experiments (n > 24 cells per sample in each experiment). The number of P-bodies per cell was significantly lower in sts5∆ orb6-as2 cells as compared to orb6-as2 cells upon Orb6 kinase inhibition relative to DMSO-treated orb6-as2 cells (P = 0.0186, Student’s t-test). No significance difference was observed when comparing orb6-as2 vs sts5∆ orb6-as2 cells treated with DMSO, P = 0.2458 (Student’s t-test). Error bars indicate SD. (C) Quantification of the experiment shown in A based on 3 independent experiments (n > 24 cells per sample in each experiment). The total P-body fluorescence intensity per cell was significantly lower in sts5∆ orb6-as2 cells as compared to orb6-as2 cells upon Orb6 kinase inhibition relative to DMSO-treated orb6-as2 cells (P = 0.0013, Student’s t-test). No significance difference was observed when comparing orb6-as2 vs sts5∆ orb6-as2 cells treated with DMSO (P = 0.1837, Student’s t-test). Error bars indicate SD. (D) mRNA levels of Sts5-regulated transcripts decrease upon Orb6-as2 kinase inhibition as compared to control, as established by qPCR analysis based on 3 independent experiments. act1 is shown as an example of a transcript that is not altered. Housekeeping genes were nda3, cdc2, and cdc22. Error bars indicate SD. (E) Ssp1-HA protein levels in control, orb6-as2, sts5Δ, and sts5Δ orb6-as2 cells cultured in the presence of 50 μM 1-NA-PP1 inhibitor in supplemented minimal medium at 25°C. Tubulin levels were determined as a loading control. (F) Quantification of Ssp1-HA/Tubulin in 4 independent experiments, as shown in E, normalized to wild-type levels. Ssp1-HA levels are significantly reduced upon Orb6-as2 kinase inhibition (P = 0.031), and are restored to wild-type levels in the sts5∆ orb6-as2 strain (P = 0.023). P values were determined using analysis of variance (ANOVA) with SPSS statistics package 22.0, followed by Games-Howell test. Error bars indicate SD.
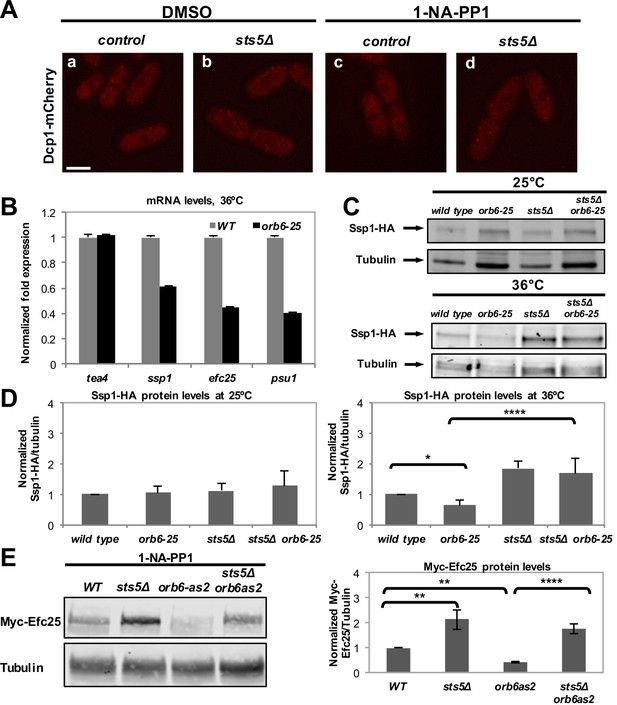
Orb6 kinase inhibits Sts5-dependent translational repression.
(A) Treatment with DMSO and 1-NA-PP1 does not induce P-body formation in control and sts5Δ cells. Dcp1-mCherry localization in control (a, c) compared with sts5Δ (b, d) cells grown in supplemented minimal medium in the presence of 50 μM 1-NA-PP1 (c and d) or DMSO (a and b) for 1 hr. Images are deconvolved projections from 12 Z-stacks separated by a step size of 0.3 μm. Bar = 5 μm. (B) Temperature-sensitive inactivation of Orb6-25 kinase promotes Sts5-dependent mRNA degradation. Decreased mRNA levels in orb6-25 cells growth at 36°C as compared with wild type as confirmed by qPCR for a selected group of mRNAs, which were identified by microarray analysis (Figure 3—source data 1). (C) Western blots showing levels of Ssp1-HA protein in orb6-25 and control cells at 25°C and at 36°C. Tubulin levels were determined as a loading control. (D) Quantification of Ssp1-HA/Tubulin ratio in the experiment shown in C, normalized to wild-type levels, based on 3 independent experiments. At the restrictive temperature (36°C), the change in Ssp1-HA level is significantly reduced in orb6-25 as compared to control cells (P = 0.044). Ssp1-HA levels are significantly higher in sts5Δ (P<0.0001) and sts5Δ orb6-25 (P<0.0001) cells as compared to orb6-25 cells at 36°C. P values were determined using analysis of variance (ANOVA) with SPSS statistics package 22.0, followed by Dunnet’s t test. Error bars denote SD. (E) Western blot showing levels of Myc-Efc25 protein in WT, sts5Δ, orb6-as2, and sts5Δ orb6-as2 strains cultured in the presence of 50 μM 1-NA-PP1 inhibitor in supplemented minimal medium at 25°C. Tubulin levels were determined as a loading control. Right panel: Quantification of Myc-Efc25/Tubulin ratio, normalized to wild-type levels, based on 3 independent experiments. Myc-Efc25 level is significantly reduced in orb6-as2 as compared to control cells (P = 0.0048). When comparing orb6-as2 with sts5Δ orb6-as2 cells, P<0.0001. Myc-Efc25 levels are significantly higher in sts5Δ (P = 0.0010) and sts5Δ orb6-as2 (P<0.0140) cells compared to wild-type cells. P values were determined using analysis of variance (ANOVA) with SPSS statistics package 22.0, followed by Tukey’s HSD post-hoc test. Error bars = SD.
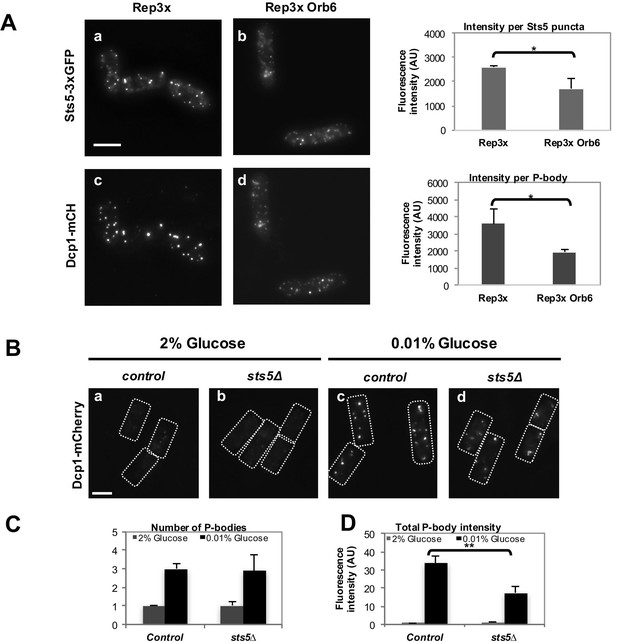
Overexpression of Orb6 inhibits Sts5 granule assembly, and Sts5 plays a role in P-body formation.
(A) Overexpression of Orb6 reduces the intensity of Sts5-3xGFP (P = 0.0227, Student’s t-test) and Dcp1-mCherry (P = 0.0352, Student’s t-test) puncta in cells cultured in supplemented minimal medium minus glucose for 1 hr.Quantification based on 3 independent experiments (N>22 cells per condition). Bar = 5 μm. (B) Dcp1-mCherry localization in control (a) compared with sts5Δ (b) cells grown in supplemented minimal medium in the presence of 0.01% glucose or 2% glucose for 1 hr. Loss of Sts5 decreases the total intensity of P-bodies induced by glucose deprivation. Bar = 5 μm. (C) Quantification of the number of P-bodies in the experiment shown in B based on 3 independent experiments relative to control cells cultured in 2% glucose. Error bars indicate SD. (N > 29 cells per condition). (D) Quantification of the total P-body intensity in the experiment shown in B based on 3 independent experiments relative to control cells cultured in 2% glucose. Error bars indicate SD. The total P-body intensity per cell was significantly lower in sts5∆ cells as compared to control cells upon 1 hr of growth in supplemented minimal medium containing 0.01% glucose relative to control cells (P = 0.0069, Student’s t-test; N > 74 cells per strain). The total P-body intensity was not significantly different among the two strains in 2% glucose (P = 0.8475, Student’s t-test; N>29 cells per strain). Error bars indicate SD.
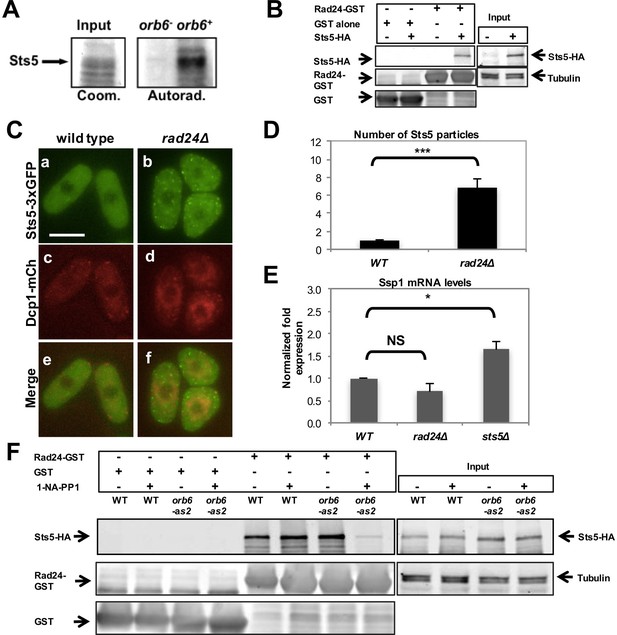
14-3-3 protein Rad24 negatively regulates Sts5 recruitment into puncta.
(A) Orb6 kinase phosphorylates Sts5 in vitro. Mob2-associated Orb6 kinase was immunoprecipitated for a kinase assay as described in the Materials and Methods and incubated with bacterially expressed Sts5 in the presence of [γ32P]ATP. (B) Endogenously expressed Sts5-HA co-purifies with bacterially expressed GST-Rad24 but not with GST alone in a pull-down assay. Three independent experiments were performed. (C) Sts5-3xGFP and Dcp1-mCherry aggregation in 2% glucose YE in WT vs rad24Δ cells. Images are deconvolved projections from 12 Z-stacks separated by a step size of 0.3 μm. Bar = 5 μm. (D) Quantification of the experiment shown in C based on 3 independent experiments (n > 27 cells per strain in each experiment). The number of Sts5 particles is significantly higher in rad24∆ relative to wild-type control cells (P = 0.0005, Student’s t-test). Error bars indicate SD. (E) qPCR analysis showing ssp1 mRNA levels are unchanged in rad24∆ cells compared with WT (P = 0.160) and increased in sts5∆ cells compared with WT (P = 0.044). When comparing sts5∆ with rad24∆ cells, P=0.006. P values were determined using analysis of variance (ANOVA) with SPSS statistics package 22.0, followed by Games-Howell post-hoc test. Housekeeping genes were nda3, act1, and cdc2. Error bars indicate SD. Three independent experiments were performed. (F) Physical association between endogenously expressed Sts5-HA and bacterially expressed GST-Rad24 is lower upon inhibition of Orb6-as2 with 50 μM 1-NA-PP1 compared with DMSO treatment (lanes 7 and 8). Sts5-HA association with GST-Rad24 remains unchanged in wild-type cells in the presence or absence of the inhibitor (lanes 5 and 6). GST-only control is shown in lanes 1–4.
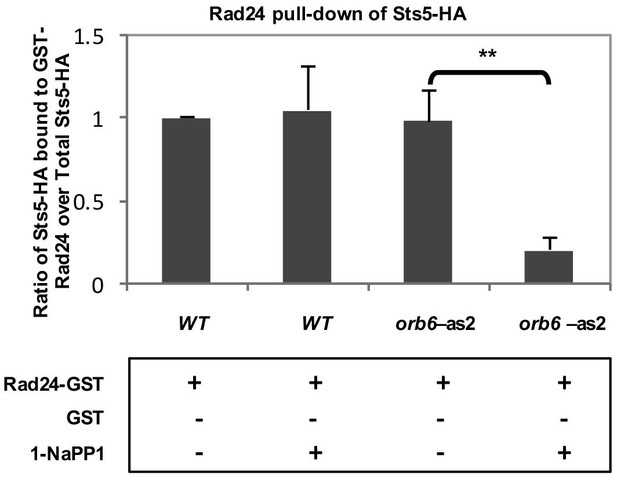
Quantification of the physical association between endogenously expressed Sts5-HA and bacterially expressed GST-Rad24.
Quantification of pull-down experiment depicted in Figure 5D showing a significant reduction (P = 0.0025; Student’s t test) in the physical interaction between endogenously expressed Sts5-HA and bacterially expressed GST-Rad24 upon inhibition of Orb6-as2 kinase with 50 μM 1-NA-PP1. Error bars denote SD. Three independent experiments were performed.
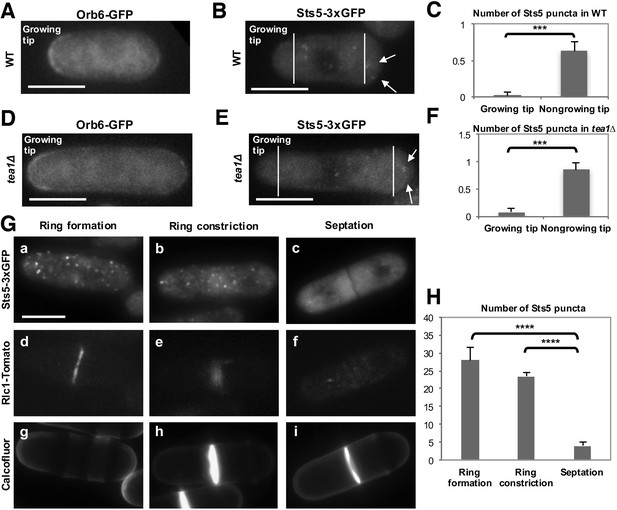
Role of Orb6 kinase in Sts5 granule assembly during the cell cycle.
(A–F) Active Orb6 kinase localization spatially anti-correlates with Sts5 recruitment into puncta in interphase cells. A. Orb6-GFP localizes to the growing cell tip in small monopolar wild-type cells. Orb6-GFP is enriched at the growing old cell end as compared to the non-growing new cell end. Bar = 5 μm. (B) Sts5-3xGFP aggregation increases towards the new cell end in monopolar wild-type cells. Images are deconvolved projections from 12 Z-stacks separated by a step size of 0.3 μm. Bar = 5 μm. (C) The average number of Sts5-3xGFP puncta per cell at the non-growing new end is significantly higher as compared to the growing old end (P<0.0001, Student’s t-test). Error bars denote SD. Three independent experiments were performed (N = 31 cells). (D) Orb6-GFP localizes to the growing cell tip in tea1∆ cells. Bar = 5 μm. (E) Sts5-3xGFP recruitment onto puncta increases towards the non-growing tip in tea1∆ cells. Images are deconvolved projections from 12 Z-stacks separated by a step size of 0.3 μm. Bar = 5 μm. (F) The average number of Sts5-3xGFP puncta per cell at the non-growing end in tea1∆ cells is significantly higher as compared to the growing old end (P<0.0009, Student’s t-test). Error bars denote SD. Three independent experiments were performed (N = 24 cells). We used calcofluor staining to identify growing tips and measured monopolar tea1∆ cells that were growing from the previous old end, which facilitated definitive identification of the nongrowing cell end. (G–H) Orb6 kinase activity temporally anti-correlates with Sts5 assembly into puncta during mitosis. (G) (a, b and c) Localization of Sts5-3xGFP in cells undergoing cell division; (d, e and f) visualization of Rlc1-Tomato; (g, h and i) calcofluor staining of cell wall and septum. Bar = 5 μm. (H) Quantification of the number of Sts5 puncta in dividing cells during cytokinetic ring formation, ring constriction, and septation. Ring formation vs septation, P<0.0001; ring constriction vs septation, P<0.0001; ring formation vs ring constriction P = 0.588 (N>20 cells per condition). P values were determined using analysis of variance (ANOVA) with SPSS statistics package 22.0, followed by Tukey’s HSD post-hoc test. Three independent experiments were performed.
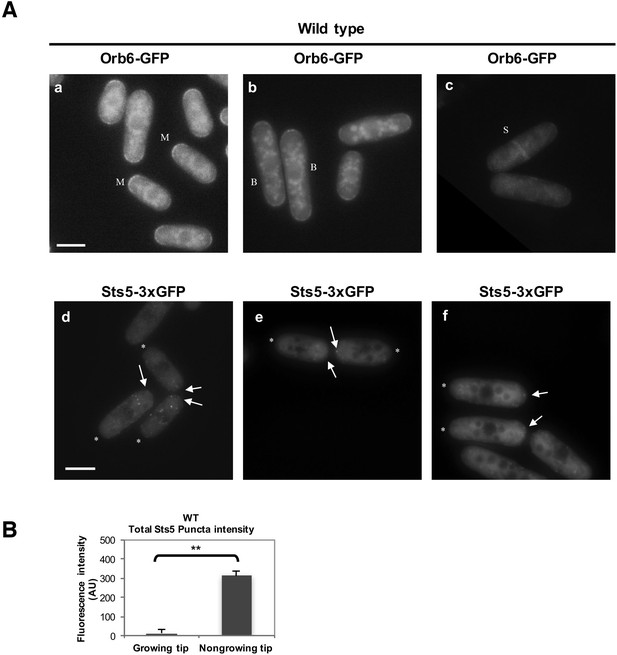
Additional images of Orb6-GFP and Sts5-3xGFP localization in monopolar WT cells and quantification of total Sts5-3xGFP granule intensity at growing and nongrowing tips.
(A) (a, b, c) Localization of Orb6-GFP in wild-type monopolar (M) (a), bipolar (B) (b), and septated (S) (c) cells cultured in supplemented minimal medium. (d, e, f) Localization of Sts5-3xGFP puncta (see arrows) in wild-type monopolar (M) cells (d, e, f). * indicates growing tip. Bar = 5 μm. (B) The average total intensity per cell of Sts5-3xGFP puncta at the non-growing new end is significantly higher as compared to the growing old end (P = 0.0059, Student’s t-test). Error bars denote SD. Three independent experiments were performed (N = 31 cells).
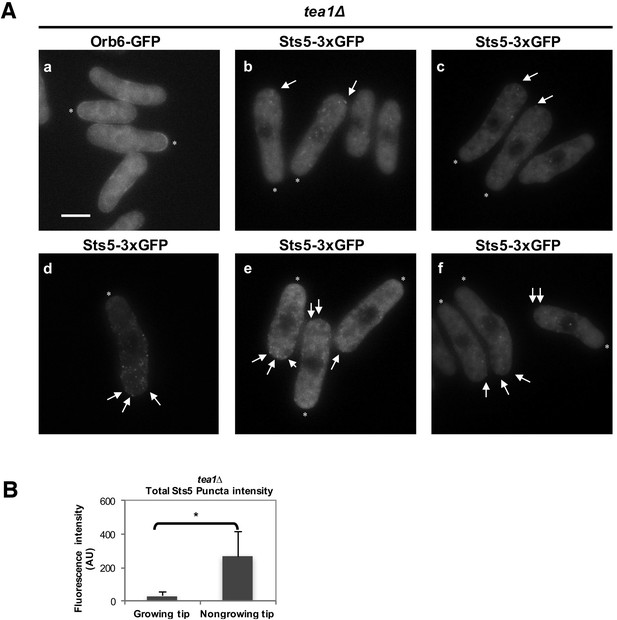
Additional images of Orb6-GFP and Sts5-3xGFP localization in monopolar tea1Δ cells and quantification of total Sts5-3xGFP granule intensity at growing and nongrowing tips.
(A) (a) Localization of Orb6-GFP in tea1Δ monopolar cells, and (b–f) localization of Sts5-3xGFP puncta (see arrows) in tea1Δ monopolar cells cultured in supplemented minimal medium. * indicates growing tip. Bar = 5 μm. (B) The average total intensity per cell of Sts5-3xGFP puncta at the non-growing end in tea1∆ cells is significantly higher as compared to the growing old end (P = 0.0435, Student’s t-test). Error bars denote SD. Three independent experiments were performed (N = 24 cells).
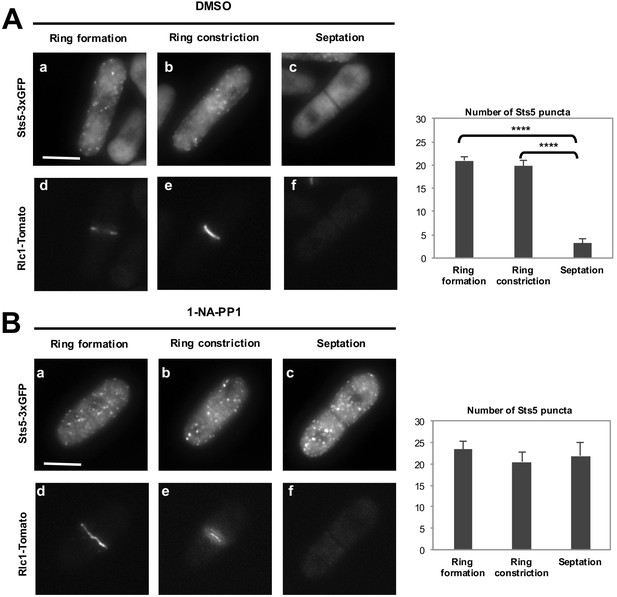
Orb6 kinase inhibition prevents the dissolution of Sts5-3xGFP puncta after completion of mitosis.
(A) (a, b, c) Localization of Sts5-3xGFP in cells undergoing cell division and (d, e, f) expressing Rlc1-Tomato during cytokinetic ring formation, ring constriction, and septation in orb6-as2 cells treated with DMSO. Bar = 5 μm. Right panel: Quantification of Sts5-3xGFP puncta in DMSO-treated orb6-as2 cells. Ring formation vs septation, P<0.0001; ring constriction vs septation, P<0.0001; ring formation vs ring constriction P = 0.345 (N = 18 cells per condition). P values were determined using analysis of variance (ANOVA) with SPSS statistics package 22.0, followed by Tukey’s HSD post-hoc test. Three independent experiments were performed. (B) (a, b, c) Localization of Sts5-3xGFP in cells undergoing cell division and (d, e, f) expressing Rlc1-Tomato during cytokinetic ring formation, ring constriction, and septation in orb6-as2 cells treated with 50 mM 1-NA-PP1 for 1 hr. Bar = 5 μm. Right panel: Quantification of Sts5-3xGFP puncta in 1-NA-PP1-treated orb6-as2 cells. Ring formation vs septation, P = 0.764; ring constriction vs septation, P<0.773; ring formation vs ring constriction P = 0.392 (N = 18 cells per condition). P values were determined using analysis of variance (ANOVA) with SPSS statistics package 22.0, followed by Tukey’s HSD post-hoc test. Three independent experiments were performed.
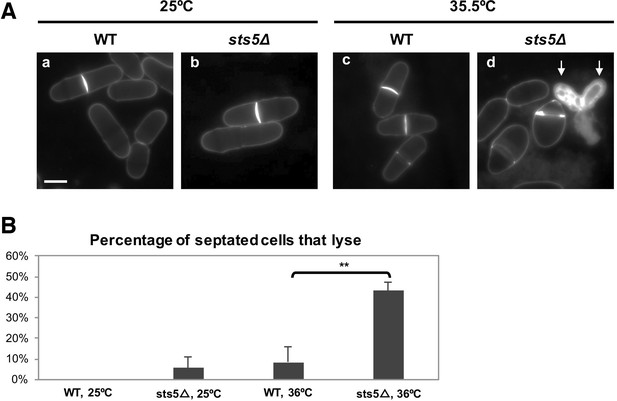
sts5∆ cells display increased cell lysis during cell separation.
(A) sts5∆ cells display increased cell lysis during cell separation (d) at the restrictive temperature (35.5°C). Arrows indicate sister cells that have lysed. (B) Quantification of the percentage of septated cells that lyse in WT and sts5∆ cells at 25°C and 35.5°C. Quantification based on 3 independent experiments. Percentage of septated cells that lyse is significantly greater in sts5Δ cells as compared to controls at 35.5°C (P = 0. 0.0022, Student’s t-test, N>176 cells per condition). When comparing WT and sts5∆ cells 25°C, P = 0.1583 (Student’s t-test).
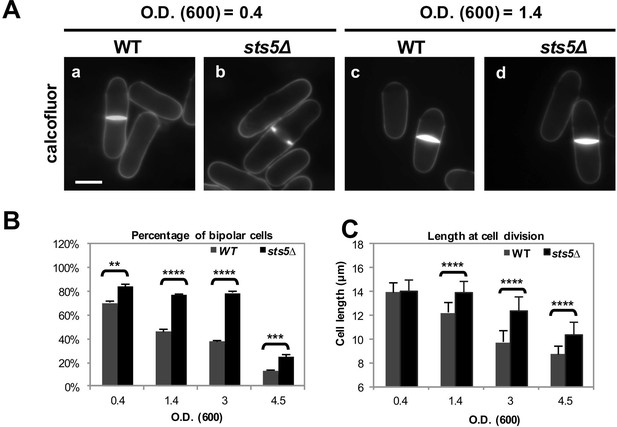
Sts5 modulates bipolar growth activation during exponential cell proliferation and during nutritional stress.
(A) sts5∆ cells display a delayed morphological response to nutritional stress induced by high cell density as compared with wild type cells. Cells were stained with calcofluor. Bar = 5 μm. (B) Quantification of the percentage of bipolar cells in control versus sts5∆ cells in the experiment depicted in A. Percentage bipolar cells was significantly higher in sts5∆ cells versus control cells during exponential growth (OD600 <0.4) (P = 0.0013, Student’s t test) and at OD600 = 1.4 (P<0.0001, Student’s t test), OD600 = 3 (P<0.0001, Student’s t test), and OD600 = 4.5 (P = 0.0003, Students’ t test). Error bars indicate SD. At least 3 independent experiments were performed (N>64 for each strain per cell density condition). Cells undergoing cell division were not included. (C) Quantification of cell size (defined as cell length at division) in control versus sts5∆ cells in the experiment depicted in A. Cell size was significantly longer in sts5∆ cells versus control at OD600 = 1.4 (P<0.0001, Student’s t test), OD600 = 3 (P<0.0001, Student’s t test), and OD600 = 4.5 (P<0.0001, Student’s t test). Error bars indicate SD. At least 3 independent experiments were performed (N>16 for each strain per cell density condition).
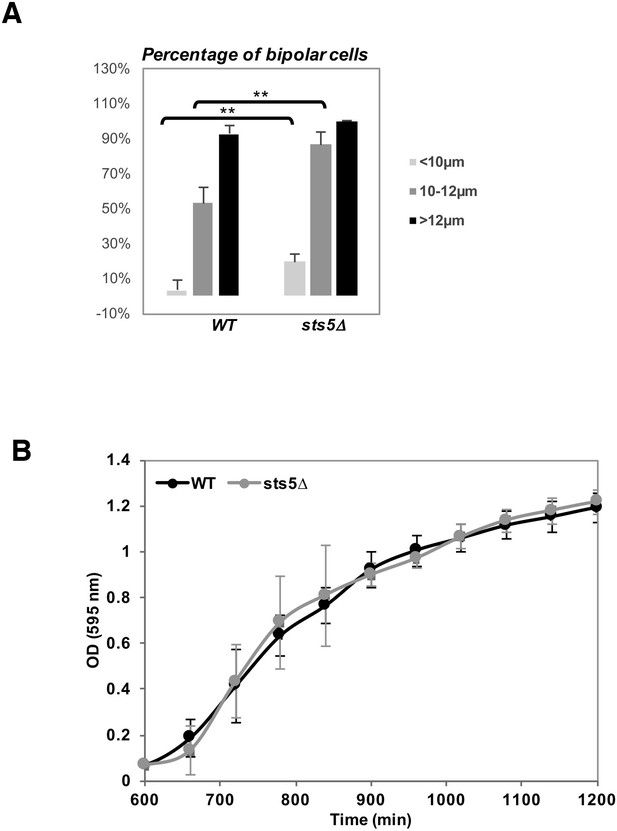
Increased bipolarity of sts5Δ vs wild-type cells is not due to changes in cell size or overall cell growth.
(A) Quantification of the percentage of bipolar cells in wild type versus sts5∆ cells comparing small cells (<10 mm in length; P = 0.0050, Student’s t-test), larger cells (10–11.99 mm in length; P = 0. 0.0012 Student’s t-test) and cell with a length longer than 12 µm cells; P = 0.0256).Quantifications based on 4 independent experiments (N>239 cells per strain). (B) Growth curves (OD 595 nm) of wild-type versus sts5∆ cells as measured by the TECAN system. Cell were grown in supplemented minimal medium.
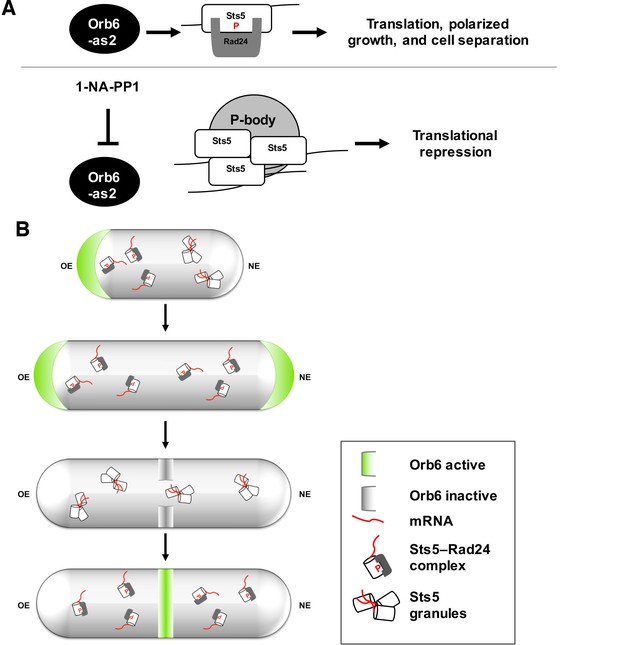
A model of spatial control of translational repression and polarized growth by Orb6 kinase and mRNA binding protein Sts5.
(A) Orb6 kinase prevents Sts5 recruitment into larger RNP granules by promoting the association between Sts5 and the 14-3-3 protein Rad24. Upon Orb6 kinase inhibition, Sts5 proteins are recruited into larger RNP granules and co-localize with P-bodies, leading to reduced mRNA levels and translational repression. (B) In small monopolar cells Orb6 kinase is localized at the growing old end. Sts5 recruitment in larger granules is observed at the new end, lacking Orb6 kinase activity. In larger bipolar cells Orb6 kinase is localized at both cell tips, and Sts5 recruitment is reduced at both cell ends and throughout the cell. During mitosis, Orb6 kinase is inactivated by the SIN pathway, which allows Sts5 recruitment into larger RNP granules and translational repression. Once cell separation is complete, Orb6 kinase activity resumes, promoting Sts5 disassembly, translational derepression, and cell separation.
Additional files
-
Supplementary file 1
List of strains used in this study.
- https://doi.org/10.7554/eLife.14216.028





