Mitochondrial genetic diversity, selection and recombination in a canine transmissible cancer
Figures
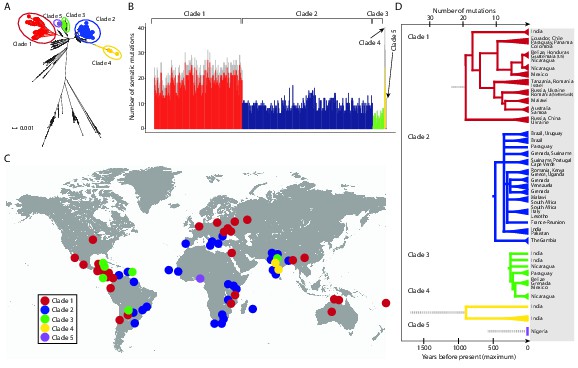
CTVT has acquired mtDNA by horizontal transfer at least five times.
(A) Maximum likelihood phylogenetic tree constructed with complete mtDNA sequences from 449 CTVT tumours and 590 dogs. Coloured and black dots represent CTVT and dog mtDNA respectively. Scale bar indicates base substitutions per site. (B) Number of somatic substitution mutations per CTVT tumour. Coloured bars indicate somatic mutations acquired by each tumour since mtDNA capture. Grey bars indicate substitutions absent from normal dog mtDNA haplotypes but common to all tumours within a clade; thus the early somatic or rare germline status of these variants is unknown. (C) Geographical distribution of clades. Coloured dots represent locations from which one or more CTVT tumours were collected. (D) Simplified representation of maximum likelihood phylogenetic trees for each clade. Trees illustrate nodes with bootstrap support >60, and shaded triangles represent coalescence of individual branches within each group. Two tumours were collected in the United States and the Netherlands respectively from dogs imported from Guatemala and Romania. Discontinuous grey lines represent contributions of substitutions absent from normal dog mtDNA haplotypes but common to all tumours within a clade. Assuming a constant accumulation of mutations within and between clades, approximate number of somatic mutations and estimated timing is shown. Maximum likelihood trees upon which these representations are based are found in Figure 1—source data 2.
-
Figure 1—source data 1
Maximum likelihood phylogenetic tree of CTVT mtDNA.
Maximum likelihood phylogenetic tree constructed using 449 complete CTVT mitochondrial genomes and 590 complete dog mitochondrial genomes. All sequences are labelled with sample identifier, country, breed and haplotype name. The sample identifier for CTVT hosts is the sample name (Supplementary file 1), the sample identifier for the publicly available dogs is the accession number. Scale bar indicates base substitutions per site.
- https://doi.org/10.7554/eLife.14552.004
-
Figure 1—source data 2
Maximum likelihood phylogenetic trees for CTVT clades 1 to 5.
Maximum likelihood phylogenetic trees for CTVT mtDNA in (A) clade 1 (n = 170) (B) clade 2 (n = 252) (C) clade 3 (n = 22) (D) clade 4 (n = 3) and (E) clade 5 (n = 2), rooted with haplotypes CTVT1 to CTVT5 respectively, which contain clade-defining germline and potential somatic substitutions specific to each clade (Figure 1—figure supplement 4). Bootstrap values were calculated from 100 bootstrap replicates and are shown where bootstrap values ≥60. Scale bars indicate base substitutions per site. Clade 5 contains only two tumours, which are identical both to each other and to the CTVT5 haplotype; thus the tree for this clade was created separately and does not have a scale bar.
- https://doi.org/10.7554/eLife.14552.005
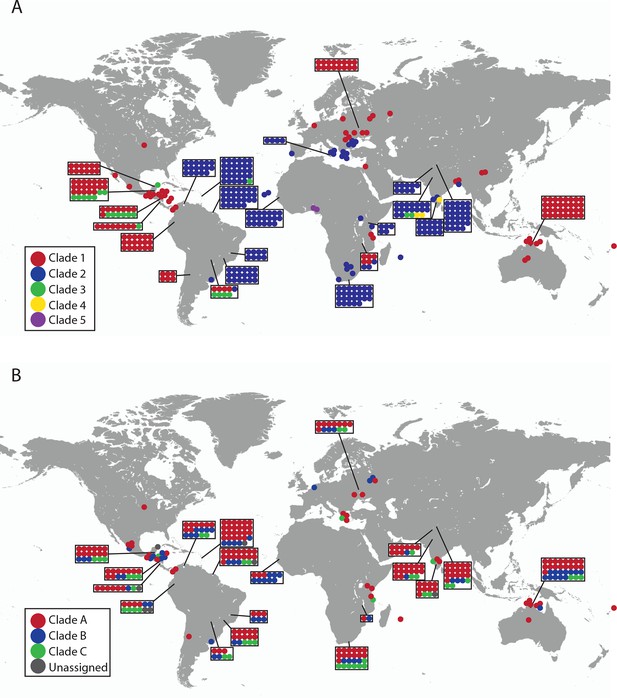
Geographical locations and mtDNA clades for CTVT tumours and hosts.
Each dot represents the location of (A) CTVT tumours, coloured by CTVT mtDNA clade; or (B) CTVT hosts, coloured by dog mtDNA clade.
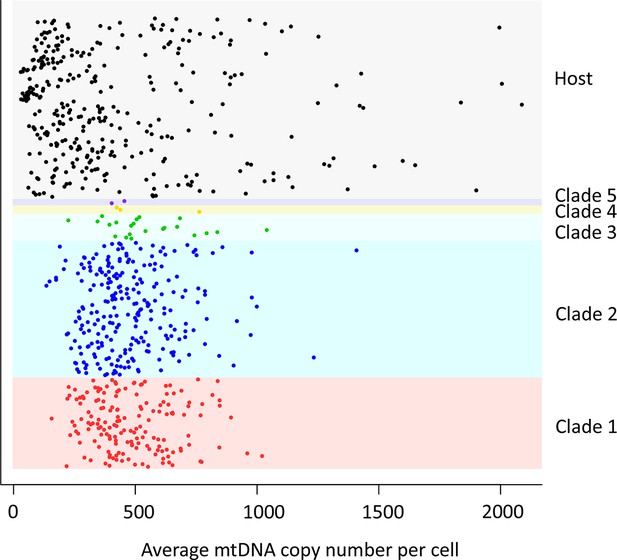
mtDNA copy number in CTVT.
MtDNA copy number was estimated by normalising mtDNA sequence coverage to whole genome sequence coverage (Supplementary file 2A). Each point represents an individual tumour (labelled by clade) or host. MtDNA copy number in tumours was not normalised for host contamination. Host and tumour samples with average MT coverage >300X (see Supplementary file 2A) were excluded from the analysis and from calculation of average number of mtDNA copies per cell.
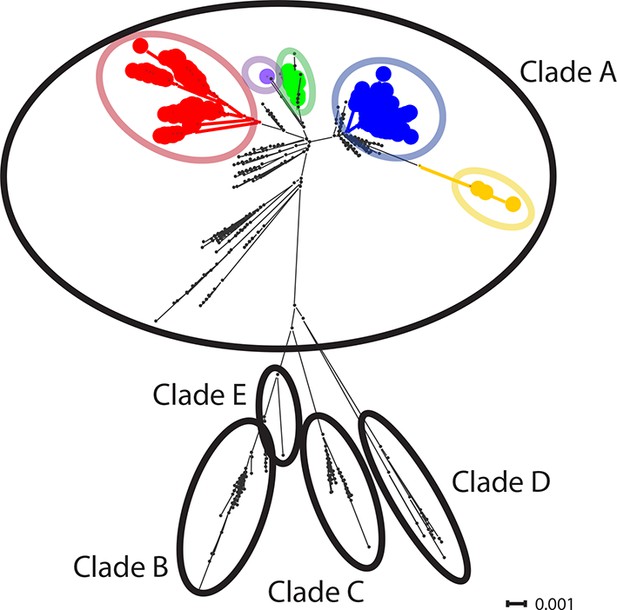
CTVT mtDNA clades 1 to 5 all arose from dog mtDNA clade A.
Maximum likelihood phylogenetic tree constructed with complete mtDNA sequences from 449 CTVT tumours and 590 dogs. Coloured and black dots represent CTVT and dog mtDNA respectively (CTVT mtDNA clade colours are represented as in Figure 1A). Dog mtDNA clades A to E are labelled (Savolainen et al., 2002; Vila et al., 1997). Scale bar indicates base substitutions per site.
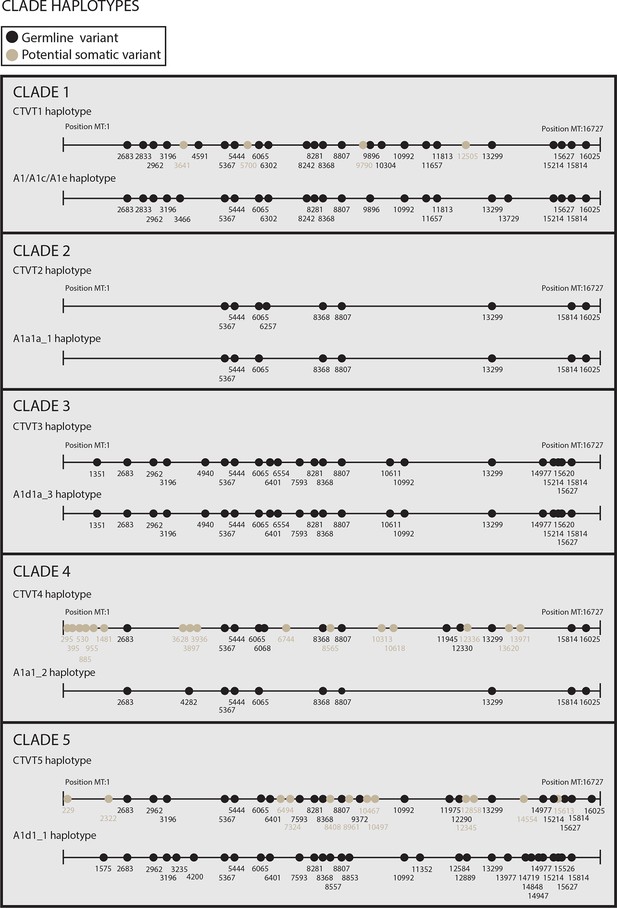
Reconstructed donor haplotypes for CTVT mtDNA clades 1 to 5.
Diagrams representing the likely donor haplotype for each of the CTVT mtDNA clades 1 to 5. The coordinates for each substitution variant position are shown, and substitutions are colour-coded either as 'germline' (i.e. they are present in all tumours within a clade and are found in the most closely related dog mtDNA haplotype, which is represented below each of the clade diagrams or they are found in the most closely related dog mtDNA haplotype only); or 'potential somatic' (i.e. they are present in all tumours within a clade but are not found in the most closely related dog mtDNA haplotype).
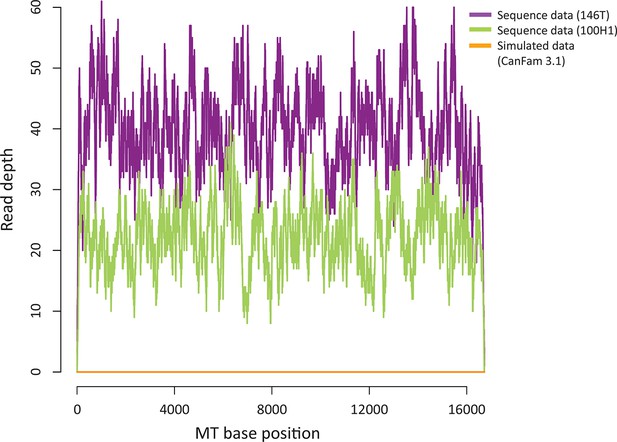
Sequence contribution of nuclear-encoded mtDNA (NuMTs).
Sequence read depth across the MT genome for a representative CTVT tumour (146T) and host (100H1) sequenced in this study to ~0.3X whole genome average coverage. This is compared with sequence read depth for simulated reads from CanFam3.1 (excluding the MT chromosome); reads were simulated to ~0.3X whole genome average coverage.
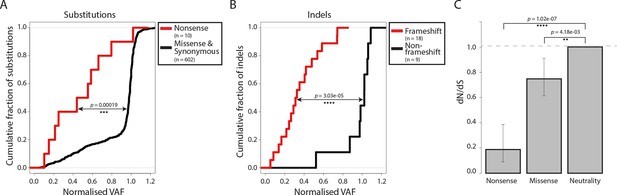
Negative selection operates to prevent the accumulation of gene-disrupting mutations in CTVT.
Cumulative distribution functions for variant allele fraction (VAF) for gene-disrupting (A) substitutions and (B) indels. P-values were calculated using two-sample Kolmogorov-Smirnov tests. (C) dN/dS for somatic nonsense and missense substitutions. P-values were calculated using a likelihood ratio test with parameters estimated using a Poisson model. Error bars indicate 95 percent confidence intervals.
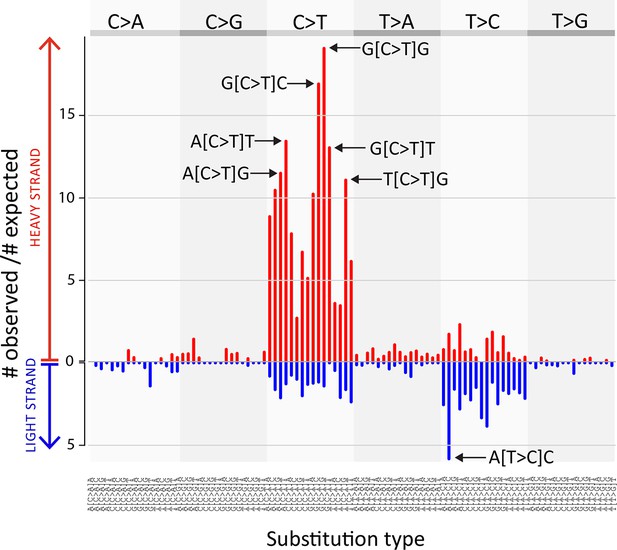
CTVT mtDNA somatic mutation spectrum.
CTVT somatic mutations displayed by mutation type (in pyrimidine context) with 5’ and 3’ context and strand. Each of 96 mutation classes is displayed on the horizontal axis, with mutations occurring on the heavy strand displayed in red on the positive axis, and light strand mutations displayed in blue on the negative axis. The normalised substitution rate represents the (number of observed)/(number of expected) mutations, given mtDNA genome triplet content. Distinctive peaks are individually labelled. Only mutations on the 'conservative somatic list' were used (see Materials and methods and Supplementary file 4C).
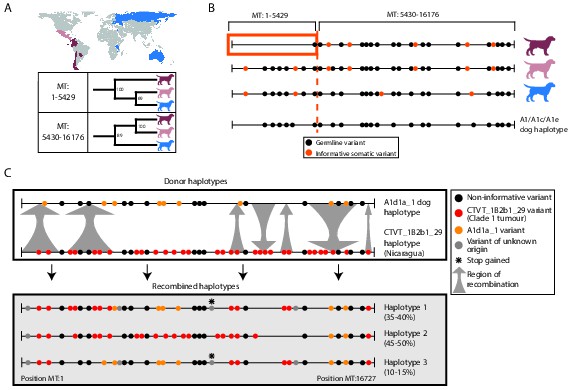
Ancient and modern mtDNA recombination in CTVT.
(A) Maximum likelihood phylogenetic trees constructed using segments MT:1–5429 and MT:5430–16176 from clade 1 CTVT mtDNAs. Three clade 1 mtDNA haplotype groups are represented by coloured dog silhouettes, and their geographical distributions are colour-coded on the map. Bootstrap values were calculated from 100 iterations. Maximum likelihood trees upon which these representations are based are found in Figure 3—source data 1. (B) Simplified haplotype diagrams for clade 1 CTVT mtDNAs derived from groups shown in (A). Germline variants were present in the donor mtDNA that founded clade 1, represented by the A1/A1c/A1e dog haplotype (see Figure 1—figure supplement 4). Region putatively replaced by recombination is outlined with orange box. (C) Recombination detected in tumour 559T (Nicaragua). The estimated per cent contribution of each recombined haplotype to the mtDNA population within 559T CTVT cells is shown, and grey arrows indicate likely sites of recombination.
-
Figure 3—source data 1
Ancient mtDNA recombination in CTVT clade 1.
Maximum likelihood cladograms constructed using clade 1 mtDNA positions (A) 1-5429bp and (B) 5430-16176bp (see Materials and methods). Trees were constructed with 153 clade 1 CTVT mtDNAs rooted with the CTVT1 haplotype, which contains clade 1 clade-defining germline and potential somatic substitutions (Materials and methods, Figure 1—figure supplement 4). Bootstrap values were calculated from 100 bootstrap replicates and are shown where bootstrap values ≥60.
- https://doi.org/10.7554/eLife.14552.014
Diagram showing phylogenetic trees constructed using PhyML and RAxML.
https://doi.org/10.7554/eLife.14552.028Additional files
-
Supplementary file 1
Sample information.
Summary of information available for 449 CTVT tumours and 338 hosts sequenced in this study. Includes data on location, year of collection, CTVT mtDNA clade, tumour and host mtDNA haplotypes, breed, age and sex.
- https://doi.org/10.7554/eLife.14552.015
-
Supplementary file 2
Sequencing coverage and tumour cell fraction.
(A) Average per-base coverage for whole genome (CanFam3.1) and for mtDNA genome (CanFam3.1; NC_002008). (B) List of 11 CTVT hosts with low coverage mtDNA regions. (C) Estimated tumour cell fraction for 449 CTVT tumours; tumour cell fraction was estimated by calculating the average VAF for variant substitutions present in tumour but not in matched host for each tumour.
- https://doi.org/10.7554/eLife.14552.016
-
Supplementary file 3
Confirmation of CTVT diagnosis.
Quantitative PCR (qPCR) was performed for LINE-MYC, a CTVT-specific rearrangement (Katzir et al., 1985; Katzir et al., 1987). Each reaction was performed in triplicate and a standard curve was used to detect relative DNA input at each locus. 'Normalised input' represents the relative LINE-MYC input detected in each sample normalised to ACTB ('Confirmation of canine transmissible venereal tumour (CTVT) diagnosis', Materials and methods). In general, we consider normalised input >0.05 as indicative of presence of LINE-MYC. Sufficient DNA was not available for samples 1380T and 1381T; diagnosis in these cases was performed with histopathology. *2T was grown as a xenograft.
- https://doi.org/10.7554/eLife.14552.017
-
Supplementary file 4
Single point substitution variant lists.
(A) Total number of substitution variants (n = 1005) identified in 449 CTVT tumours. (B) CTVT tumour somatic substitutions list (n = 928), including the average VAF value normalised for host contamination (see Materials and methods 'Classification of tumour substitutions'). Back mutations are not included on the list. (C) CTVT tumour conservative somatic substitutions list (n = 835), including the average VAF value normalised for host contamination (see Materials and methods 'Classification of tumour substitutions'). Back mutations are not included on the list. (D) Germline clade defining substitutions lists. Substitutions present in the pool of host substitutions and also shared between all samples within a clade (see Materials and methods 'Classification of tumour substitutions'). (E) Potential somatic substitutions lists. Substitutions not present in the pool of host substitutions, but shared between all samples within a clade (see Materials and methods 'Classification on tumour substitutions'). (F) Total number of substitution variants (n = 1152) identified in 338 CTVT host samples and 252 publicly available dog mitochondrial genomes (see Supplementary file 8).
- https://doi.org/10.7554/eLife.14552.018
-
Supplementary file 5
Summary of small insertions and deletions (indels).
(A) Total number of insertions and deletions identified in tumours (n = 27), including the average VAF value normalised for host contamination (see Materials and methods) (B) Total number of homoplasmic insertions and deletions in CTVT hosts (n = 7), including the average VAF value (see Materials and methods).
- https://doi.org/10.7554/eLife.14552.019
-
Supplementary file 6
Annotation of single point substitutions.
Annotation of individual point substitution mutations in (A) 449 CTVT tumours (see list Supplementary file 4A, excluding back mutations) and (B) 338 CTVT hosts (see list Supplementary file 4F). Annotation was performed using Variant Effect Predictor (McLaren et al., 2010). In cases where a single substitution affects two different genes, the two annotations are shown on different lines.
- https://doi.org/10.7554/eLife.14552.020
-
Supplementary file 7
Annotation of insertions and deletions (indels).
Annotation of individual indels (A) unique to CTVT tumours and (B) homoplasmic in CTVT hosts. Annotation was performed using Variant Effect Predictor (McLaren et al., 2010).
- https://doi.org/10.7554/eLife.14552.021
-
Supplementary file 8
Publicly available mitochondrial dog genomes used in the study.
Summary of Genbank accession numbers and metadata for 252 publicly available dog mitochondrial genomes included in this study (see Materials and methods 'Extracting substitution variants from publicly available dog sequences' and Supplementary file 4F).
- https://doi.org/10.7554/eLife.14552.022
-
Supplementary file 9
Timing analysis.
Methods used to estimate the time since the origin of CTVT clades 1 to 5. Potential somatic substitutions (see Materials and methods 'Classification of tumour substitutions' and Supplementary file 4E) are those which are shared between all tumours within a clade, but that are not found in the normal dog population; thus we cannot confirm their germline or early somatic status. The upper panel lists time estimates (years before present for the origin of each clade) assuming potential somatic substitutions are somatic and arose after mtDNA horizontal transfer; the lower panel lists time estimates (years before present for the origin of each clade) assuming potential somatic substitutions are germline and were originally present on the mtDNA haplotype that founded the clade.
- https://doi.org/10.7554/eLife.14552.023
-
Supplementary file 10
Summary of back mutations.
List of back mutations attributable and non-attributable to a putative recombination event.
- https://doi.org/10.7554/eLife.14552.024
-
Supplementary file 11
CTVT tumour and host mtDNA haplotype lists.
- https://doi.org/10.7554/eLife.14552.025
-
Supplementary file 12
Substitutions with corresponding VAF (before normalisation) for each of 449 CTVT tumours and 338 CTVT hosts.
Supplementary file 12 is included as accompanying zip file.
- https://doi.org/10.7554/eLife.14552.026
-
Supplementary file 13
Indels with corresponding VAF (before normalisation) for each of 438 CTVT tumours and 334 CTVT hosts.
Samples with very high coverage of the mitochondrial genome were excluded from the indel analysis (see Materials and methods 'Indel calling-Extraction and filtering'). Supplementary file 13 is included as accompanying zip file.
- https://doi.org/10.7554/eLife.14552.027





