A long non-coding RNA targets microRNA miR-34a to regulate colon cancer stem cell asymmetric division
Figures
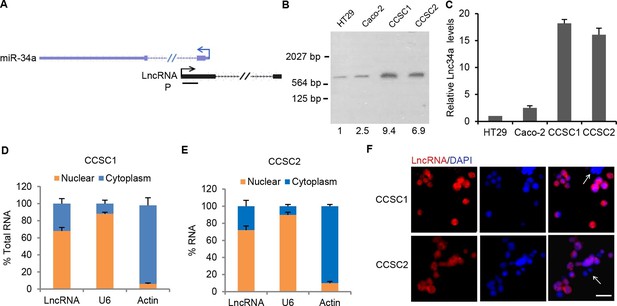
Characterization of Lnc34a.
(A) Schematic illustration of Lnc34a (shown in black) and miR-34a (shown in blue) gene structure. Lnc34a and miR-34a contain two exons and are transcribed in different directions. P, probe for Northern blot in (B). (B) Northern blot detection of Lnc34a with the probe shown in (A), quantified by Image J. (C) RT-qPCR detection of Lnc34a expression in colon cancer stem cells (CCSC1 and CCSC2) and well-established colon cancer cell lines (HT29 and Caco-2). (D, E) RT-qPCR detection of Lnc34A level in cellular fractions from CCSC1 (D) and CCSC2 (E) sphere cells. U6 and actin are the nuclear and cytoplasm controls, respectively. (F) Lnc34a expression in CCSC sphere cells detected by RNA-FISH. Scale bar, 20 μm.
-
Figure 1—source data 1
Information of CRC patients.
Tissue specimens collected from the listed CRC patients were used for analyses of Lnc34a and miR-34a expression and miR-34a promoter methylation.
- https://doi.org/10.7554/eLife.14620.004
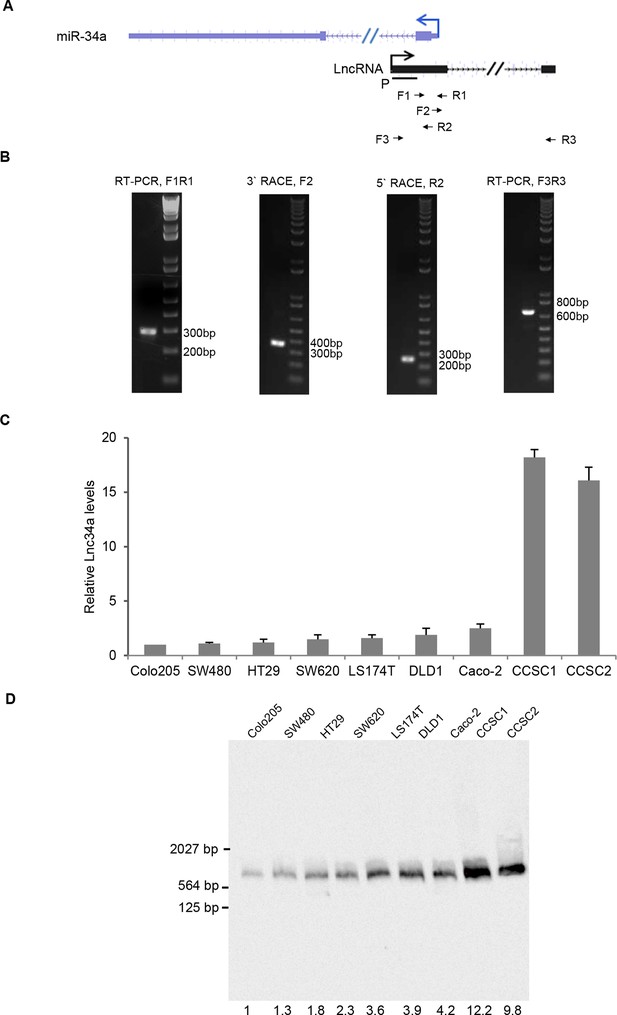
Identification of Lnc34a.
(A) Schematic illustration of Lnc34a and miR-34a gene structures. Primers for RT-PCR and rapid amplification of cDNA ends (RACE) were shown. (B) RT-qPCR and RACE that amplified Lnc34a. (C) RT-qPCR detection of Lnc34a expression in CRC cell lines. (D) Northern blot detection of Lnc34a in CRC cell lines and CCSCs. The quantification of each band was carried out using Image J.
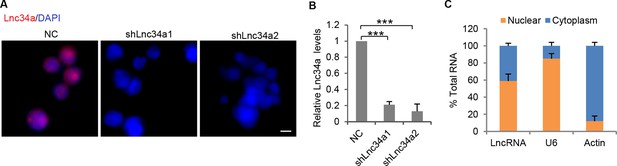
RNA FISH specificity and Lnc34a knockdown efficiency.
(A) Knockdown of Lnc34a abolished RNA FISH signals. (B) RT-qPCR showing Lnc34a knockdown efficiencies by shLnc34a1 and shLnc34a2. (C) RT-qPCR detection of Lnc34a level in cellular fractions from CCSC1with Lnc34a ectopic expression.
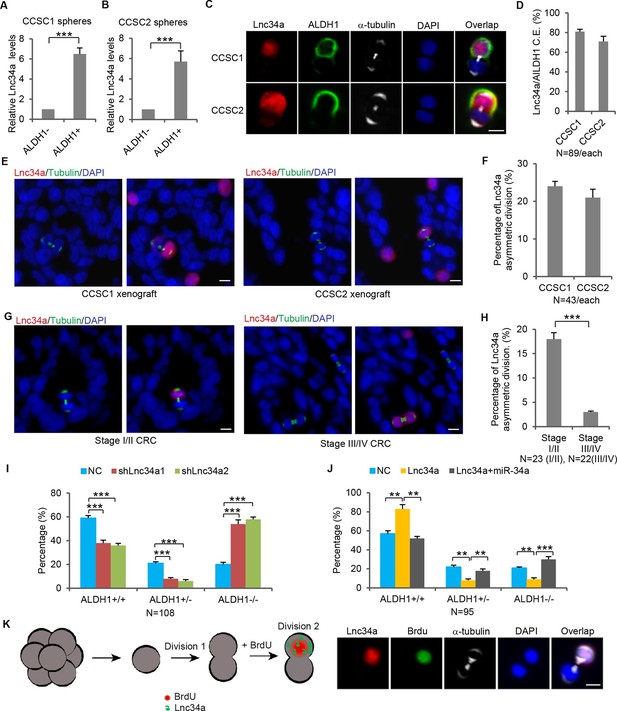
Lnc34a Asymmetry in CCSC division.
(A, B) RT-qPCR detection of lnc34a in ALDH1+ and ALDH1- populations isolated from spheres of two independent patient-derived lines, CCSC1 (A) and CCSC2 (B). Lnc34a is high in ALDH1+ (CCSC) but low in ALDH1- (non-CCSC) cells. (C) Representative images of Lnc34a distribution in dividing pairs. α-tubulin staining is consistent with the telophase (final phase of mitosis) configuration of microtubules – the midbody at the division plane during cytokinesis and asters at the poles. ALDH1 identifies the CCSC daughter. (D) Quantification of Lnc34a/ALDH1 co-expression (C.E.) in daughter compartments of dividing pairs as shown in (C). (E) Representative images of Lnc34a asymmetry in dividing pairs in xenograft tumors derived from CCSC1 and CCSC2. Dividing pairs are identified by tubulin staining. (F) Percentage of Lnc34a asymmetry in dividing pairs in CCSC xenografts as shown in (E). (G) Representative images of asymmetric and symmetric Lnc34a distribution in dividing pairs in early- and late-stage human CRC specimens. (H) Percentage of Lnc34a asymmetry in dividing pairs in human CRC specimens. (I) Effect of Lnc34a knockdown on mode of division based on ALDH1 staining of dividing cell pairs. Lnc34a knockdown decreased asymmetric (ALDH1+/ALDH1-) division and symmetric self-renewal (ALDH1+/ALDH1+), and increased differentiation (ALDH1-/ALDH-). (J) Effect of ectopic Lnc34a expression on mode of division. Ectopic Lnc34a increased symmetric self-renewal (ALDH1+/ALDH+), and reduced asymmetric division (ALDH1+/ALDH1-) and differentiation (ALDH1-/ALDH1-). The effect of ectopic Lnc34a expression was abrogated by ectopic miR-34a expression. (K) Pair-cell BrdU incorporation assay showing asymmetric proliferative potential. Left, schematic representation of the experimental approach. Single sphere cells were allowed to divide once (1st division). Cells were then treated with BrdU for 3 hr to label cells that were re-entering the 2nd division. Right, representative images showing that the Lnc34a+ cells were more proliferative and incorporated BrdU. Scale bar, 8 μm. Error bars denote s.d. of triplicates. *p<0.05; **p<0.01; ***p<0.001. p-value was calculated based on Student’s t-test.
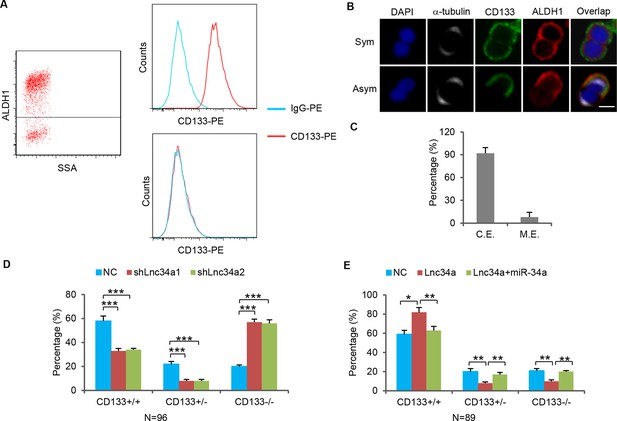
CCSCs co-express ALDH1 and CD133.
(A) FACS showing ALDH1+ sphere cells are CD133+. (B) Co-immunofluorescence of ALDH1 and CD133 showing ALDH1 and CD133 are expressed in the same daughter cell during CCSC division. (C) Percentages of CCSC divisions wherein miR-34a and ALDH1 are coexpressed (C.E.) or mutually exclusive (M.E.). (D, E) Effect of Lnc34a knockdown (D) and ectopic Lnc34a and miR-34a expression (E) on mode of division based on CD133 staining of dividing cell pairs.
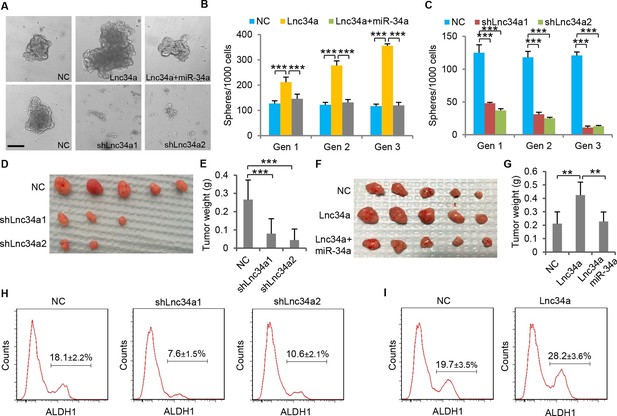
Lnc34a promotes CCSC self-renewal and tumor formation.
(A) Representative images of CCSC spheres with Lnc34a knockdown (shLnc34a1 and shLnc34a2), ectopic Lnc34a expression (Lnc34a), and ectopic Lnc34a/miR-34a expression. (B, C) Sphere formation during serial passages after Lnc34a knockdown (B) and ectopic Lnc34a and miR-34a expression (C). Equal number of cells was passaged for 3 generations to form spheres. (D, E) Knockdown of Lnc34a (shLnc34a1 and shLnc34a2) reduced tumorigenicity, shown by images (D) and weights of xenograft tumors (E). (F, G) Ectopic Lnc34a expression (Lnc34a) enhances tumorigenicity, which can be abrogated by ectopic miR-34a expression. (H, I) FACS plots identifying ALDH1+ (CCSC) populations in xenograft tumors with Lnc34a knockdown (H) or ectopic Lnc34a expression (I). Scale bar, 50 μm. Error bars denote s.d. of triplicates. **p<0.01; ***p<0.001. p-value was calculated based on Student’s t-test.
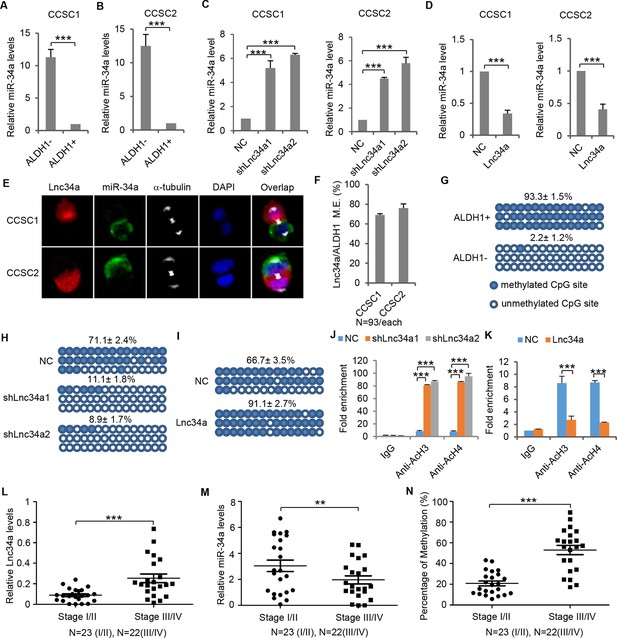
Lnc34a epigenetically silences miR-34a promoter.
(A, B) RT-qPCR of miR-34a levels in CCSC1 (A) and CCSC2 (B). (C, D) RT-qPCR of miR-34a levels in CCSC1 (C) and CCSC2 (D) spheres with Lnc34a knockdown (shLnc34a1 and shLnc34a2) or ectopic expression (Lnc34a). NC is the control vector. (E) Representative images of Lnc34a and miR-34a asymmetry in CCSC1 and CCSC2 sphere cells. (F) Quantification of (E). Lnc34a and miR-34a distributions are mutually exclusive (M.E.) during most CCSC divisions. (G) Bisulfite sequencing analysis showing miR-34a promoter methylation status in ALDH1+ (CCSC) and ALDH1- (non-CCSC) cells isolated from sphere cells. PCR products amplified from bisulfite-treated genomic DNA were cloned and sequenced to reveal the methylation status of individual CpG sites. Percentages of the methylated CpG sites (filled circles) among all scored sites are indicated. (H) Lnc34a knockdown decreased miR-34a promoter methylation in sphere cells. (I) Ectopic Lnc34a expression increased miR-34a promoter methylation in sphere cells. (J, K) ChIP-qPCR with antibodies against acetylated histones H3 and H4. Lnc34a knockdown decreased miR-34a promoter acetylation (J), while ectopic Lnc34a expression increased acetylation (K). (L) RT-qPCR measurements of Lnc34a expression in early- and late-stage CRC specimens. (M) RT-qPCR measurements of miR-34a expression in early- and late-stage CRC specimens. (N) Bisulfite sequencing analysis of miR-34a promoter methylation status in early- and late-stage CRC specimens. Scale bar, 8 μm. Error bars denote s.d. of triplicates. **p<0.01; ***p<0.001. p-value was calculated based on Student’s t-test.
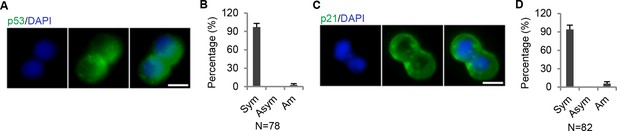
p53 symmetry.
(A) (A) Representative immunofluorescence images showing symmetric distribution of p53 during CCSC division. (B) Percentage of p53 division type. (C) Representative immunofluorescence images showing symmetric distribution of p21 during CCSC division. (D) Percentage of p21 division type. Sym, symmetric segregation; Asym, asymmetric segregation; Am, ambiguous.
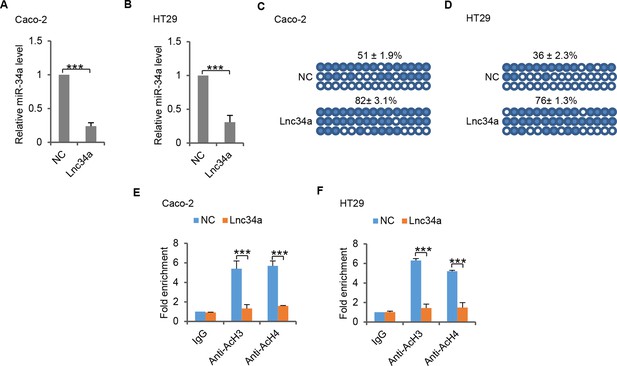
Lnc34a epigenetically silences miR-34a promoters in Caco-2 and HT29 cells.
(A, B) RT-qPCR of miR-34a levels in CRC lines Caco-2 (A) and HT29 (B). Ectopic Lnc34a expression suppressed miR-34a expression. (C, D) Bisulfite sequencing analysis showing ectopic Lnc34a expression increased miR-34a promoter methylation in Caco-2 (C) and HT29 (D). (E, F) ChIP-qPCR with antibodies against acetylated histones H3 and H4. Ectopic Lnc34a expression decreased miR-34a promoter acetylation in Caco-2 (E) and HT29 (F).
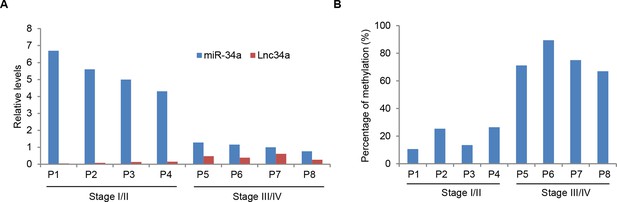
Lnc34a, miR-34a, and promoter methylation levels in CRC specimens.
(A) RT-qPCR showing Lnc34a and miR-34a expression in individual CRC specimens. Levels are normalized to corresponding actin levels. (B) Bisulfite sequencing analysis of miR-34a promoter methylation in the same CRC specimens shown in (A).
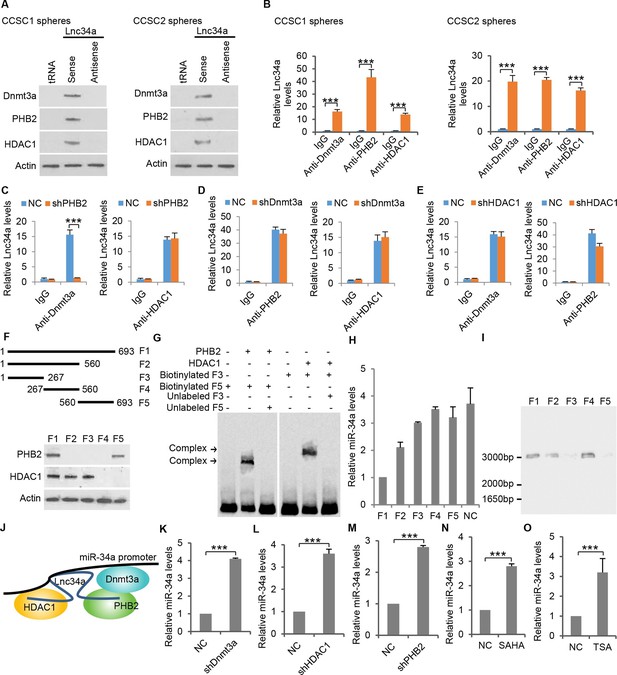
Lnc34a recruits epigenetic regulators.
(A) Western blot following RNA-pull down showing Lnc34a interaction with PHB2, Dnmt3a and HDAC1 in CCSC1 (left) and CCSC2 (right) sphere cells. RNA-pull down was performed using CCSC lysates with biotin-labeled Lnc34a, antisense and tRNA. Actin was used for input control. (B) RNA immunoprecipitation (RIP) showing Lnc34a interaction with PHB2, Dnmt3a and HDAC1 in CCSC1 (left) and CCSC2 (right) sphere cells. (C) RIP showing PHB2 knockdown disrupts Lnc34a interaction with Dnmt3a, but has no effect on Lnc34a interaction with HDAC1. (D) RIP showing Dnmt3a knockdown does not affect Lnc34a interaction with PHB2 or HDAC1. (E) RIP showing HDAC1 knockdown has limited effect on Lnc34a interaction with PHB2 or Dnmt3a. (F) Mapping PHB2 and HDAC1 interaction domains on Lnc34a. Upper panel, schematic illustration of full-length Lnc34a and the truncated fragments for RNA put-down. Lower panel, Western blot of PHB2 and HDAC1 from RNA put-down of the fragments. (G) EMSA showing Lnc34a/PHB2 (left) and Lnc34a/HDAC1 (right) interactions. (H) RT-qPCR of miR-34a levels after expressing full-length or truncated fragments of Lnc34a. (I) In vitro interaction assay binding of the truncated fragment (267–560 bp) to the DNA containing the miR-34a promoter sequence. (J) Schematic illustration of Lnc34a interaction with PHB2, Dnmt3a and HDAC1. (K, L, M) RT-qPCR showing knockdown of Dnmt3a (K), HDAC1 (L), and PHB2 (M) increased miR-34a expression in sphere cells. (N, O) RT-qPCR showing treatments with HDAC inhibitor SAHA (N) or TSA (O) increased miR-34a expression in sphere cells. Error bars denote s.d. of triplicates. ***p<0.001. p-value was calculated based on Student’s t-test.
-
Figure 5—source data 1
Potential Lnc34a-associated proteins identified by biotinylated Lnc34a pull-down and mass spectrometry.
- https://doi.org/10.7554/eLife.14620.015
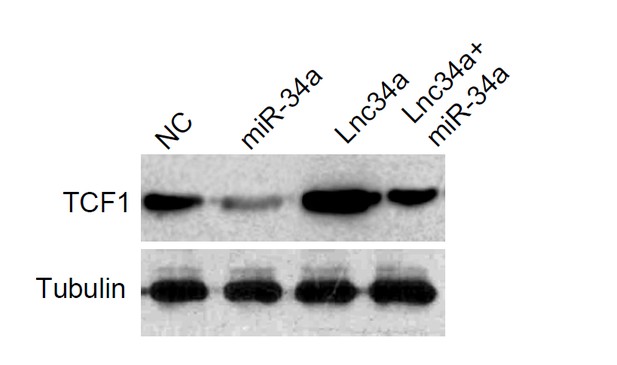
Lnc34a upregulates TCF1 via suppression of miR-34a.
Western blot showing TCF1 was suppressed by ectopic miR-34a expression and upregulated by ectopic Lnc34a expression in CCSCs. Ectopic miR-34a abrogated the upregulation of Lnc34a on TCF1.
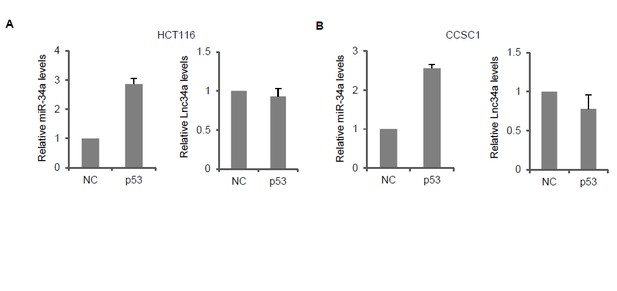
Lnc34a and induction of miR-34a by p53 does not affect each other.
(A and B) RT-qPCR showing miR-34a and Lnc34a expression level in HCT116 (A) and CCSC1 (B) cells with p53 ectopic expression.





