Premature polyadenylation of MAGI3 produces a dominantly-acting oncogene in human breast cancer
Figures
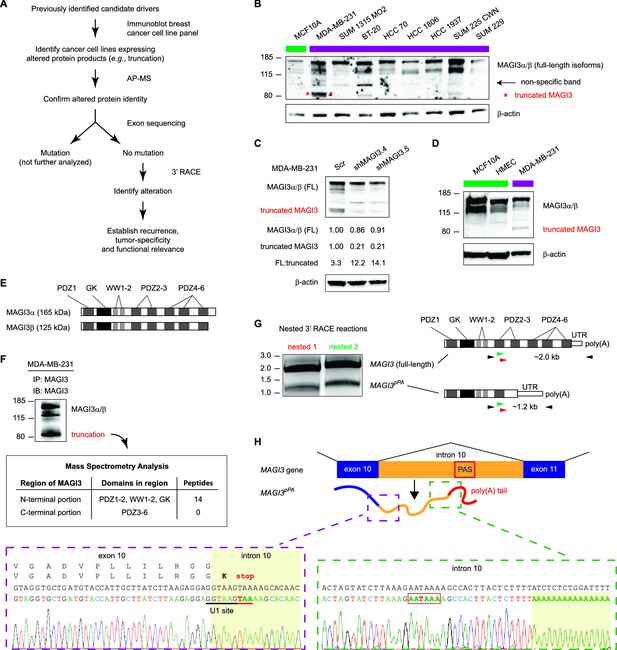
Premature polyadenylation of MAGI3 in the MDA-MB-231 breast cancer cell line causes the expression of a truncated MAGI3 protein.
(A) Strategy for identifying protein products altered by previously overlooked mechanisms. Protein products of previously identified candidate driver genes (Ni et al., 2013) were screened by immunoblotting. Altered products were identified by electrophoretic mobility shift and confirmed by affinity purification-mass spectrometry (AP-MS). Exons of corresponding genes were sequenced. Genes not affected by coding-region DNA mutations were analyzed by 3′ RACE and sequencing to identify the nature of the altered product and the associated mechanism of alteration. Alteration events caused by mechanisms not widely appreciated in cancer were subsequently investigated for tumor-specific recurrence and functional relevance. (B) Immunoblot of MAGI3 gene products in the screening panel of human breast cell lines. Non-transformed (green) and transformed (violet) cell lines are indicated. A truncated MAGI3 protein (indicated by red asterisks) is expressed in MDA-MB-231 cells. Immunoblot of β-actin is included to show loading. Approximate molecular mass markers are indicated in kDa. (C) Immunoblots of MAGI3 gene products and β-actin levels in MDA-MB-231 cells transduced with the indicated shRNA constructs. The relative levels of full-length and truncated MAGI3 proteins following RNAi-mediated depletion are normalized to β-actin levels. The ratios of full-length to truncated MAGI3 (FL:truncated) are also quantified. (D) Immunoblot of MAGI3 gene products in the indicated non-transformed cell lines (green) and MDA-MB-231 breast cancer cell line (violet). Immunoblot of β-actin is included to show loading. Approximate molecular mass markers are indicated in kDa. (E) Diagrams of the full-length MAGI3 protein isoforms α and β. Both full-length isoforms possess the indicated arrangement of 6 PDZ domains (dark grey), 2 WW domains (light grey) and a catalytically inactive GK domain with homology to the yeast guanylate kinase (black). In addition, isoform α has an extended C-terminal sequence with no known function. (F) Immunoblot for three MAGI3 protein isoforms immunoprecipitated from MDA-MB-231 cell lysate. The gel slice containing the smallest, truncated MAGI3 protein was analyzed by mass spectrometry, and peptides were mapped exclusively to the N-terminal half of the protein. Approximate molecular mass markers are indicated in kDa. (G) Detection of full-length and truncated MAGI3 mRNA isoforms by 3′ RACE. Products from two independent nested 3′ RACE reactions performed on MDA-MB-231 RNA were separated by agarose gel electrophoresis. Truncated and full-length MAGI3 transcripts are indicated. Diagrams show regions targeted for amplification and locations of primary (black) and nested (green/red) primers used for 3′ RACE. (H) Diagram of the truncated and prematurely polyadenylated MAGI3 mRNA (MAGI3pPA) generated from the MAGI3 gene locus despite the absence of genetic mutations within the gene. Green box: 3′ RACE sequencing results of MAGI3pPA show premature cleavage and polyadenylation (shaded yellow) following a cryptic PAS (outlined in red) in intron 10. The corresponding intron 10 genomic sequence is shown above in black. Violet box: Upstream of the pPA event, 3′ RACE sequencing results show a short extension of the open-reading frame into intron 10. The corresponding exon 10 and intron 10 genomic sequence is shown above in black, with the intact U1 snRNA binding site (GGTAAG) underlined in black. Intron 10 is shaded yellow. A stop codon (red underscore) occurs 6 nt following the exon-intron junction. Encoded amino acid sequence is shown in black above the nucleotide sequence (wild-type MAGI3, upper line; MAGI3pPA, lower line).
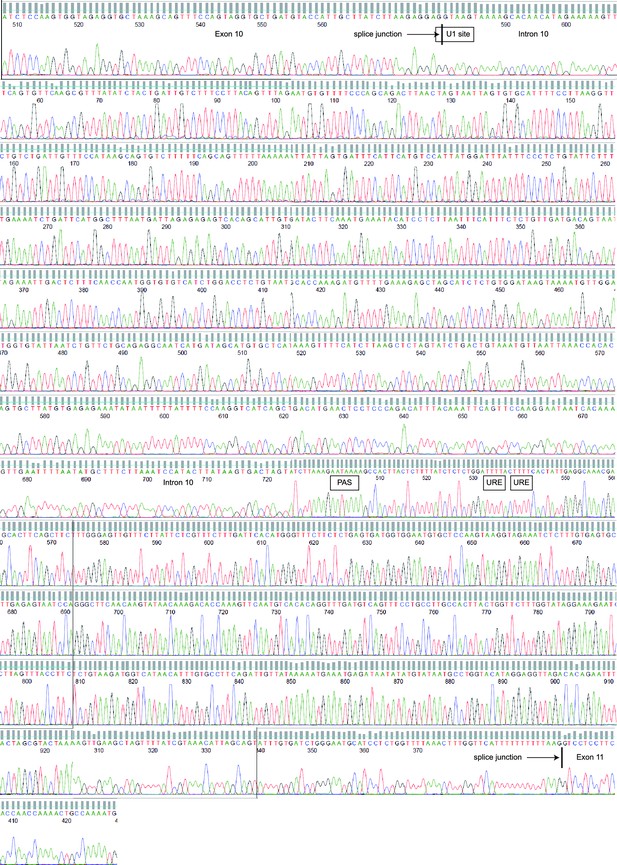
Sequencing chromatograms of MDA-MB-231 genomic DNA focusing on MAGI3 exon 10, intron 10 and exon 11.
Sequencing traces for MAGI3 exon 10, intron 10 and exon 11 are shown, with major cis-acting sequence features responsible for proper splicing and repression of premature polyadenylation annotated: splice junctions, U1 snRNA binding site and U-rich elements (URE). The location of the cryptic PAS is also indicated. No mutations were detected in these sequences.
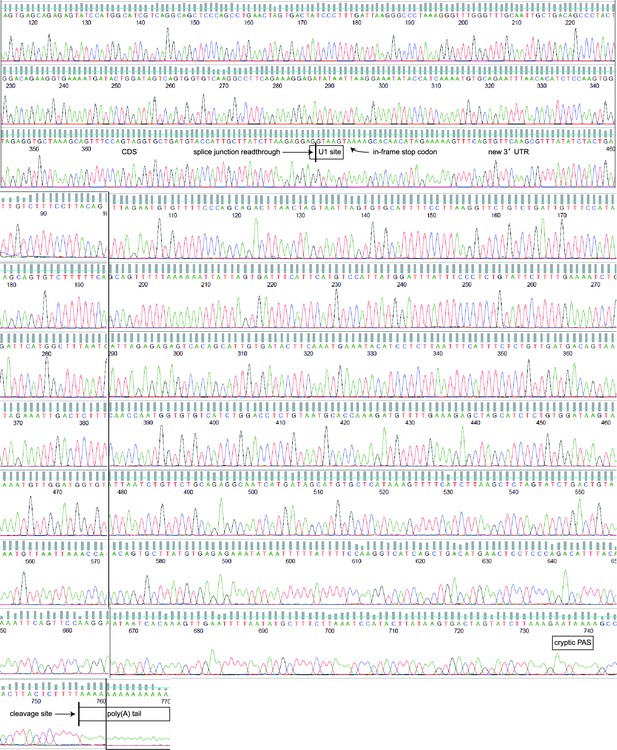
Sequencing chromatograms of the MAGI3pPA product from exon 10 to the poly(A) tail downstream of the cryptic PAS in intron 10.
Sequencing traces for the MAGI3pPA isoform are shown, with major cis-acting sequences responsible for proper splicing and repression of premature polyadenylation annotated: splice junction location and U1 snRNA binding site. The cryptic PAS, downstream cleavage site and poly(A) tail are also annotated. No mutations were detected in any of these sequences.
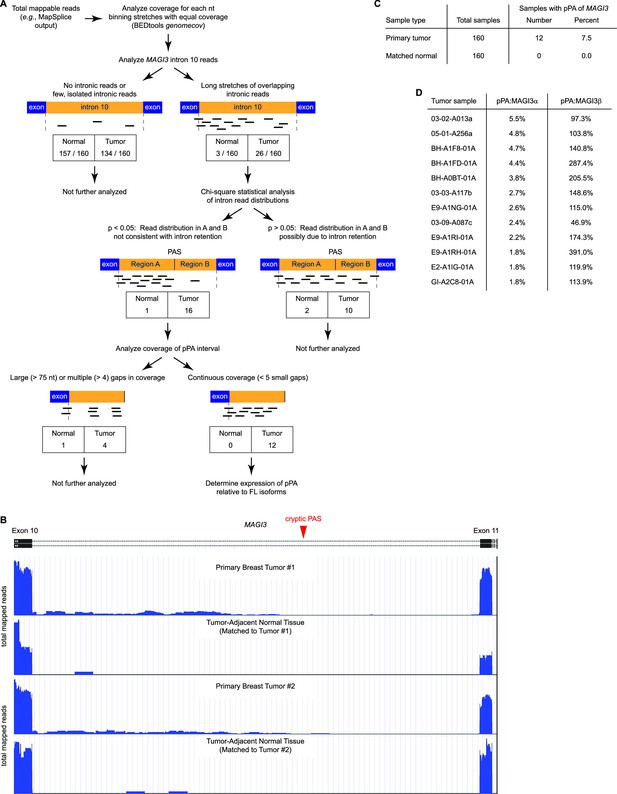
Premature polyadenylation of MAGI3 is a recurrent, tumor-specific event in breast cancer.
(A) Summary of RNA-Seq analysis, statistical tests, and thresholds used to identify primary tumor and matched, tumor-adjacent normal samples expressing MAGI3pPA. Genome coverage of total mappable reads were quantified for each sample, and read coverage across the tenth intron of MAGI3 was analyzed. Samples with sustained stretches of overlapping reads were identified and statistically analyzed for read patterns consistent with intron retention using a χ2 test. Samples with read patterns in which the intron retention null hypothesis was rejected (p<0.05) were further analyzed for continuous read coverage across the pPA-specific region. Samples with large gaps (>75 nt) or multiple gaps (>4 gaps) failed to pass the threshold set for MAGI3pPA. (B) Total read coverage (blue) of MAGI3 intron 10 in two primary breast tumors and matched, tumor-adjacent normal samples. The location of the cryptic PAS is indicated by the red arrowhead. (C) Table summary of the percentage of primary breast cancers or matched normal tissues in which premature polyadenylation of MAGI3 occurs. (D) Relative expression levels of MAGI3pPA versus full-length MAGI3α and MAGI3β isoforms in primary breast tumor samples.
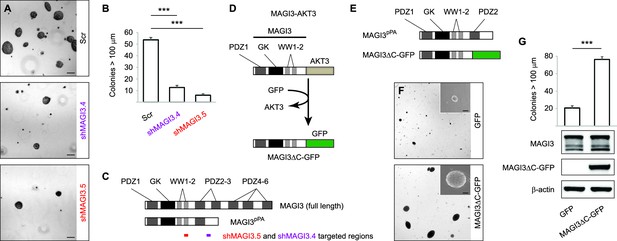
MAGI3pPA and a pPA-independent, breast cancer-associated MAGI3 truncation promote anchorage-independent growth.
(A) Anchorage-independent growth assays for MDA-MB-231 cells expressing Scr control or two independent shMAGI3 (shMAGI3.4 and shMAGI3.5) sequences that preferentially deplete MAGI3pPA. Images show representative soft agar fields for the indicated cell lines after two weeks. Scale bar represents 100 µm. (B) Quantification of soft agar colonies in the indicated cell lines (n = 3 biological replicates per cell line) after two weeks. Data are presented as mean ± SEM. ***p<0.001 (two-tailed Student’s t-tests). (C) Diagrams of full-length MAGI3 and MAGI3pPA showing the regions targeted by shMAGI3.4 and shMAGI3.5. (D) Diagrams of the protein domains of a breast cancer-associated MAGI3 truncation, fused to GFP (MAGI3ΔC-GFP), corresponding to the MAGI3-AKT3 product caused by a chromosome 1 inversion event identified by whole-genome sequencing (Banerji et al., 2012). (E) Diagrams of the protein domains of the MAGI3pPA and MAGI3ΔC-GFP truncations. (F) Anchorage-independent growth assays for MCF10A-SV40 cells expressing MAGI3ΔC-GFP or GFP control. Images show representative soft agar fields and insets show commonly observed colony sizes for the indicated cell lines after three weeks. Scale bar represents 100 µm. (G) Quantification of transformed colonies in the indicated cell lines (n = 3 biological replicates per cell line) after three weeks (upper panel). Immunoblots of MAGI3ΔC-GFP, full-length MAGI3 isoforms, and β-actin levels in the cell lines (lower panels). Data are presented as mean ± SEM. ***p<0.001 (two-tailed Student’s t-test).
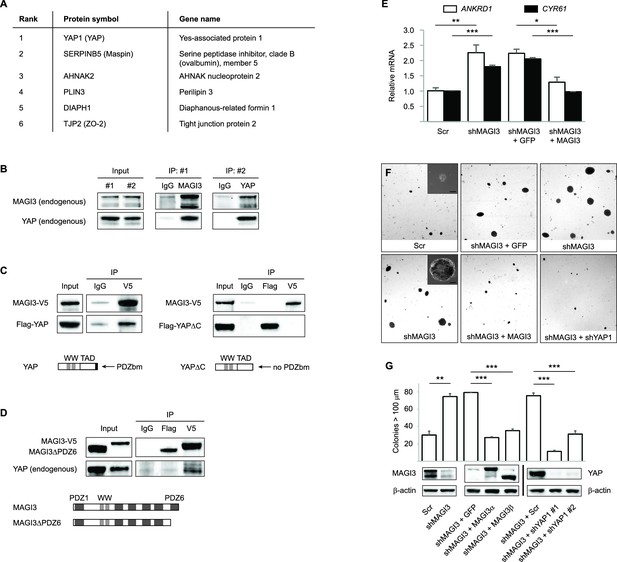
MAGI3 interacts with the YAP oncoprotein and suppresses YAP-dependent transformation.
(A) List of the top candidate MAGI3-interacting proteins in MCF10A cells as identified by AP-MS. (B) Endogenous MAGI3/YAP interaction in MCF10A cells by co-IP. Immunoblots of MAGI3 and YAP from input and indicated IP lysates are shown. (C) Immunoblots of full-length MAGI3-V5 and either full-length Flag-YAP (left) or mutant Flag-YAPΔC (right), in which the C-terminal YAP PDZ-binding motif (FLTWL) is deleted, from the indicated input and IP lysates (HEK293T cells). (D) Immunoblots of full-length YAP and either full-length MAGI3-V5 or PDZ6-deleted MAGI3-V5 from the indicated input and IP lysates (HEK293T cells). (E) The relative expression of YAP target genes ANKRD1 and CYR61 in MCF10A cells, as determined by qPCR, in cells expressing shMAGI3 or non-silencing Scr control, with or without MAGI3 cDNA or non-rescuing GFP control (n = 3 technical replicates per cell line). Data are presented as mean ± SEM. *p<0.05, **p<0.01, ***p<0.001 (two-tailed Student’s t-tests). (F) Anchorage-independent growth assays for MCF10A-SV40 cells expressing the indicated shRNA and/or cDNA constructs. Images show representative soft agar fields and insets show commonly observed colony sizes for the indicated cell lines after three weeks. Scale bar represents 100 µm. (G) Quantification of transformed colonies in the indicated cell lines (n = 3 biological replicates per cell line) after three weeks (upper panel). Immunoblots of full-length MAGI3 isoforms, YAP and β-actin levels in the cell lines (lower panels). Data are presented as mean ± SEM. **p<0.01, ***p<0.001 (two-tailed Student’s t-tests).
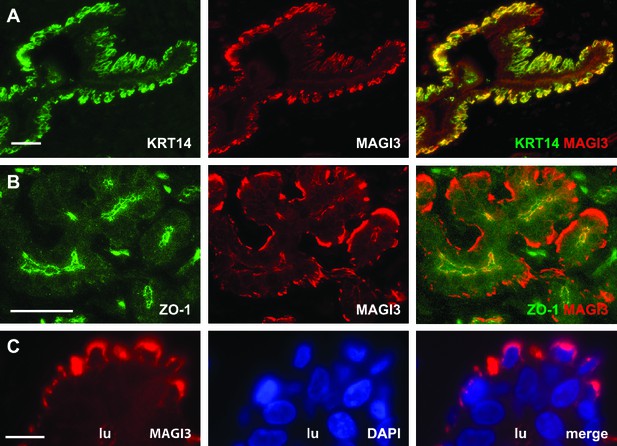
MAGI3 expression and localization in normal human mammary epithelial cells.
(A) Immunofluorescence of reduction mammoplasty tissue showing MAGI3 (red) expression and colocalization with the basal epithelial cell marker, KRT14 (green). Scale bar represents 50 µm. (B) Immunofluorescence of reduction mammoplasty tissue showing MAGI3 (red) colocalization with the tight junction protein ZO-1 (green) in human mammary luminal cells. Scale bar represents 50 µm. (C) Immunofluorescence of reduction mammoplasty and high magnification of MAGI3 (red) localization in the cytoplasm of basal/myoepithelial cells. DAPI (blue) was used to visualize nuclei. Scale bar represents 10 µm.
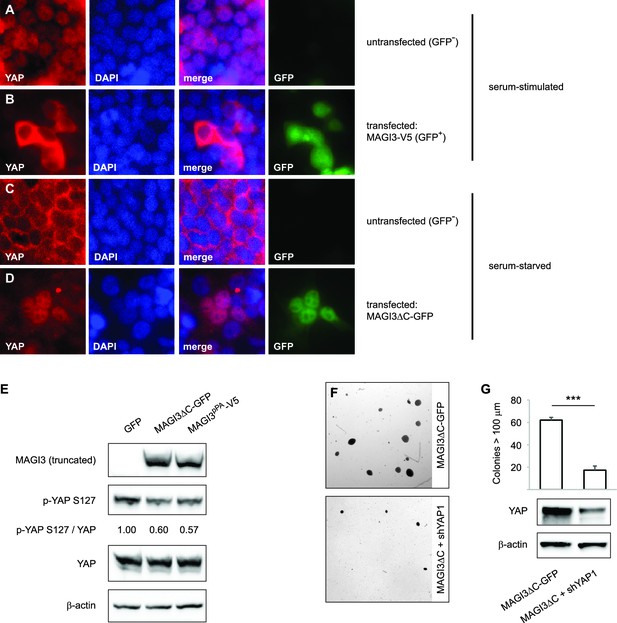
Expression of truncated MAGI3 leads to increased YAP activation and YAP-dependent transformation.
(A-B) Immunofluorescence of serum-stimulated HEK293T cells showing YAP (red) localization. GFP positivity indicates full-length MAGI3-transfected cells. DAPI (blue) was used to visualize nuclei. (A) Field containing predominantly untransfected cells is shown. (B) Field containing MAGI3-transfected cells is shown. (C-D) Immunofluorescence of serum-deprived HEK293T cells showing YAP (red) localization. GFP indicates expression and localization of MAGI3ΔC-GFP. DAPI (blue) was used to visualize nuclei. (C) Field containing predominantly untransfected cells is shown. (D) Field containing MAGI3ΔC-GFP-transfected cells is shown. (E) Immunoblots of V5-tagged MAGI3pPA, MAGI3ΔC-GFP, Ser127-phosphorylated YAP, total YAP and β-actin levels in HEK293T cells transfected as indicated. The relative levels of Ser127-phosphorylated YAP in the indicated lysates are normalized to total YAP levels. (F) Anchorage-independent growth assays for MCF10A-SV40 cells expressing MAGI3ΔC-GFP with or without shYAP1. Images show representative soft agar fields for the indicated cell lines after three weeks. Scale bar represents 100 µm. (G) Quantification of transformed colonies in the indicated cell lines (n = 3 biological replicates per cell line) after three weeks (upper panel). Immunoblots of YAP and β-actin levels in the cell lines (lower panels). Data are presented as mean ± SEM. ***p<0.001 (two-tailed Student’s t-tests).
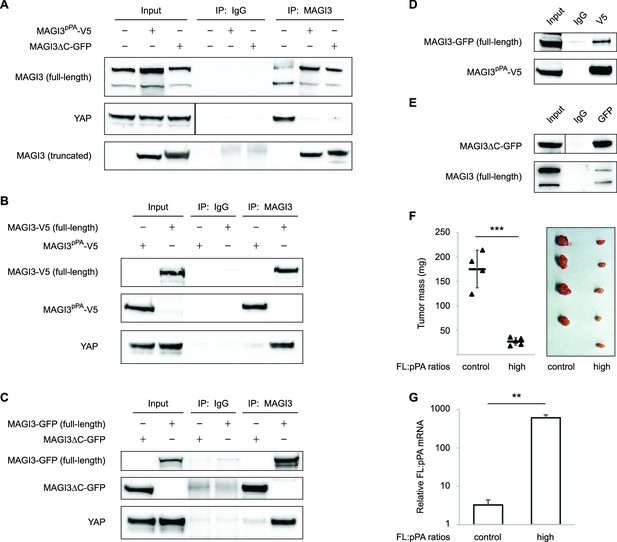
Dominant-negative MAGI3 truncations interact with full-length MAGI3 proteins to interfere with MAGI3/YAP interaction.
(A) Immunoblots of full-length endogenous MAGI3, endogenous YAP, V5-tagged MAGI3pPA and MAGI3ΔC-GFP from the indicated input and IP lysates (HEK293T cells). (B) Immunoblots of V5-tagged MAGI3pPA, full-length MAGI3-V5 and endogenous YAP from the indicated input and IP lysates (HEK293T cells). (C) Immunoblots of MAGI3ΔC-GFP, full-length MAGI3-GFP and endogenous YAP from the indicated input and IP lysates (HEK293T cells). (D) Immunoblots of full-length MAGI3-GFP and V5-tagged MAGI3pPA from the indicated input and IP lysates (HEK293T cells). (E) Immunoblots of MAGI3ΔC-GFP and endogenous full-length MAGI3 from the indicated input and IP lysates (HEK293T cells). (F) Quantification of tumor mass (left panel) and photograph (right panel) of MDA-MB-231 cells grown as orthotopic xenografts in NOD/SCID mice. Tumors with high ratios of FL:pPA MAGI3 (n = 5 mice) or unmodified control ratios (n = 4 mice) are indicated. Data are plotted as individual data points along with mean ± SD. *** p<0.001 (two-tailed Student’s t-tests). (G) The relative expression ratios of FL:pPA mRNA in control- (n = 4) or high-ratio (n = 5) MDA-MB-231 tumor xenografts, as determined by qPCR. Data are presented as mean ± SEM. ** p<0.01 (two-tailed Student’s t-test).
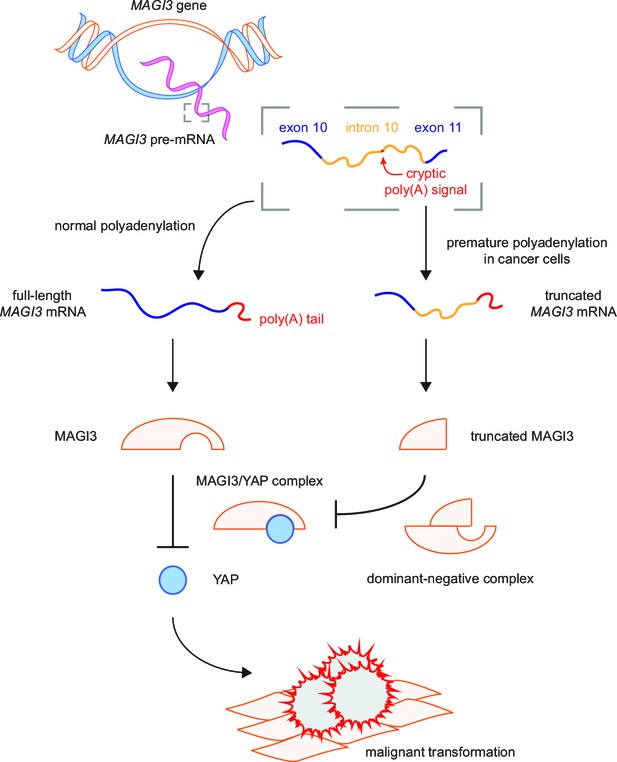
Model for the mechanism by which premature polyadenylation of MAGI3 contributes to malignant transformation.
In normal cells, the proper post-transcriptional processing of transcripts produced by the MAGI3 gene locus gives rise to full-length MAGI3 proteins that negatively regulate the YAP oncoprotein by forming an inhibitory complex. In cancer cells, premature polyadenylation of MAGI3 results in the production of truncated MAGI3 proteins in addition to full-length MAGI3. Truncated MAGI3 is unable to bind YAP but can interact with full-length MAGI3. This dominant-negative interaction may prevent full-length MAGI3 from forming an inhibitory complex with YAP, thus resulting in YAP-mediated malignant transformation.
Tables
Primer sequences used for 3′ RACE.
| Name | Primer Sequence (5′ to 3′) |
|---|---|
| M3Q2-F | CTGTGTCCTCGGTCACACTC |
| M3-F12 | TATCCATGGCATCGTCAGGC |
| M3-F13 | GTTGCTGCTACCCCTGTCAT |
| UAP-R | GGCCACGCGTCGACTAGTAC |
Primer sequences used for qPCR.
| Gene | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) |
|---|---|---|
| ANKRD1 | AGTAGAGGAACTGGTCACTGG | TGGGCTAGAAGTGTCTTCAGAT |
| CYR61 | GGTCAAAGTTACCGGGCAGT | GGAGGCATCGAATCCCAGC |
| GAPDH | CCATGGGGAAGGTGAAGGTC | TAAAAGCAGCCCTGGTGACC |
| MAGI3 (FL) | AGCAGTTTCCAGTAGGTGCT | TGTCGTCCTCGGGTTGTTTT |
| MAGI3 (pPA) | AGCAGTTTCCAGTAGGTGCT | GAGGTCCAGATGACACACCA |
Antibodies used in this study.
| Antibody | Host | Vendor/catalog number | IB | IP | IF |
|---|---|---|---|---|---|
| β-actin | Mouse | Abcam (Cambridge, MA) ab6276 | |||
| Flag (HRP-) | Rabbit | Cell Signaling Technology (Danvers, MA) 2044 | X | ||
| Flag | Mouse | Sigma-Aldrich F3165 | X | ||
| GFP | Rabbit | Abcam ab290 | X | X | |
| KRT14 | Mouse | Vector Laboratories (Burlingame, CA) VP-C410 | X | ||
| MAGI3 | Mouse | Abcam ab11509 | X | ||
| MAGI3 | Rabbit | Novus Biologicals (Littleton, CO) NBP2-17210 | X | X | X |
| mouse (Alexa488-) | Goat | Thermo Fisher Scientific A11001 | X | ||
| mouse (HRP-) | Goat | Cell Signaling Technology 7076 | X | ||
| normal IgG | Mouse | Santa Cruz Biotechnology (Dallas, TX) sc-2025 | X | ||
| normal IgG | Rabbit | Santa Cruz Biotechnology sc-2027 | X | ||
| rabbit (Alexa546-) | Goat | Thermo Fisher Scientific A11010 | X | ||
| rabbit (HRP-) | Goat | Cell Signaling Technology 7074 | X | ||
| V5 (HRP-) | Rabbit | Abcam ab1325 | X | ||
| V5 | Mouse | Thermo Fisher Scientific R960 | X | X | |
| YAP | Mouse | Santa Cruz Biotechnology sc-101199 | X | X | X |
| YAP | Rabbit | Santa Cruz Biotechnology sc-15047 | X | X | |
| p-YAP Ser 127 | Rabbit | Cell Signaling Technology 4911 | X | ||
| ZO-1 | Mouse | T 339100 | X |
Additional files
-
Supplementary file 1
Comprehensive list of candidate MAGI3-interacting proteins in MCF10A cells.
- https://doi.org/10.7554/eLife.14730.016
-
Supplementary file 2
Top human PDZ domains predicted to bind the YAP PDZ-binding motif.
- https://doi.org/10.7554/eLife.14730.017





