Genetic dissection of the Transcription Factor code controlling serial specification of muscle identities in Drosophila
Figures
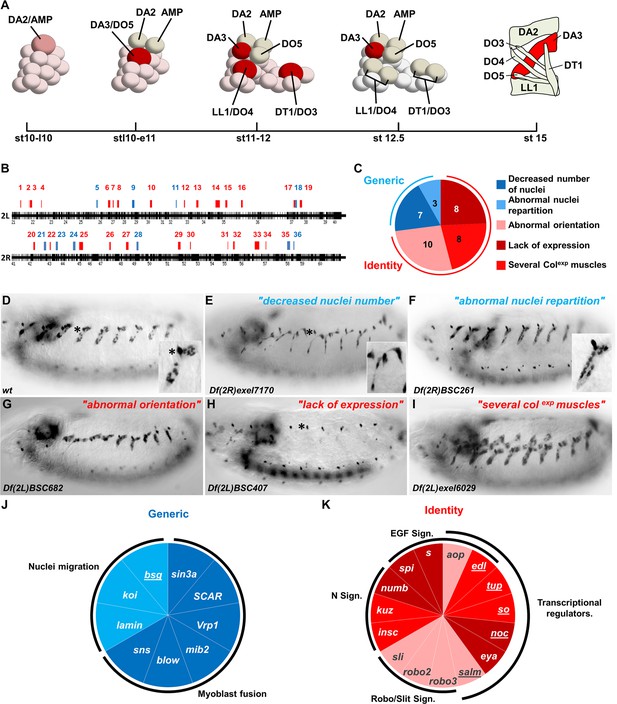
Genetic identification of muscle identity genes.
(A) Diagrammatic representation of the sequential emergence of four PCs (large cells) from the Col expressing PMC, followed by PC into FC divisions (embryonic stages (st) 10–12.5) and the corresponding muscle pattern at stage 15. The name of each PC, FC and muscle is indicated. Col expression is in red, color intensity indicating expression level. (B) Gene density along chromosome 2L and 2R, schematized by black bars. Position and size of each of 36 regions identified in our screen are indicated by red or blue bars. (C) Pie chart showing repartition of the DA3 phenotypes into two classes of generic (blue), and identity (red) defects. (D–I) Col immunostaining of late stage 15 embryos; (D) wt and (E–I), representative examples (deficiency name indicated) of each phenotypic class. The asterisk in (D,E,H) labels a dorsal class IV md neuron expressing Col. In this, and following figures, lateral views of embryos are shown, anterior to the left. (J,K) Pie charts associating individual genes with generic myogenic (K) or identity (L) mutant phenotypes. See also Figure 1—source data 1 for phenotypes.
-
Figure 1—source data 1
36 chromosomal deficiencies showing DA3 muscle phenotypes.
Numbering indicates the position of the deleted region along the chromosome, schematized in Figure 1B. The phenotypes observed at embryonic stage 16 were classified as either identity or generic muscle defects, and ranked in different types (Figure 1C). Identified genes responsible for the deficiency phenotype are indicated together with their known or predicted biochemical function.
- https://doi.org/10.7554/eLife.14979.003
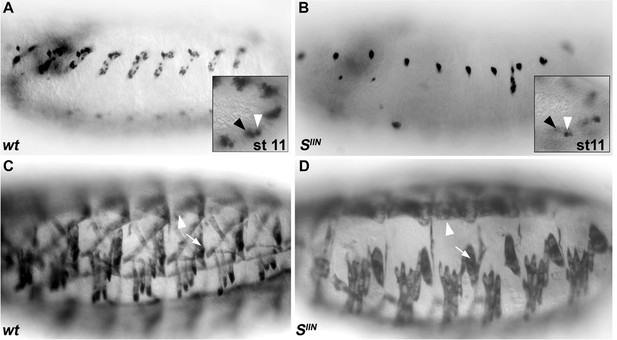
EGF-R signaling is required for a normal pattern of DL muscles.
(A,B) wt DA3 Col expression at stage 16 (A), is lost in star (SIIN) mutant embryos (B). Insets show Col expression in the DA3/DO5 (white arrowhead) and DT1/DO3 (black arrowhead) PCs (stage 11). Col expression is only detected in the posterior-most DT1/DO3 PC in SIIN mutants (B). (C–D) MHC staining of wt (C) and SIIN (D) embryos; the DA2 is present (arrowhead in D compare to C) in SIIN embryos, while the only DL muscle forming in absence of EGF-R signaling is DT1 (arrow in D, compare to C). Of note, muscles issued from dorsal PCs, including the alary muscles, form.
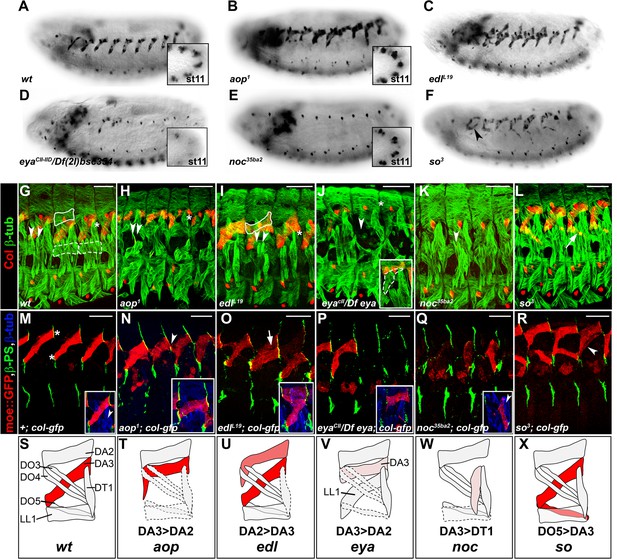
Specific muscle patterning defects in aop, edl, eya, noc and so mutant embryos.
(A–F) Late stage 15 embryos stained for Col, to visualize the DA3 muscle. (A) wt, (B–F) embryos homozygous mutant for aop, edl, eya, noc and so null alleles with their names indicated. Inserts in (A,B,D,E) show Col expression in PCs, stage 11. (G–L) stage 16 embryos stained for Col (red) and β3-tubulin (green) to visualize all body wall muscles; arrowheads point to LT1 and LT2, asterisks indicate DT1; DA2 is surrounded by a line in G,I, and LL1 by a dotted line in G. (G) wt embryo. (H) aop1 (I) edll19; Col is expressed in DA2 and DA3. (J) eyaCII/Df(2L)BSC354; DA3 Col expression is lost; inset, LL1>DA3 transformation. (K) noc35ba2: Col expression is lost. (L) so3; Col expression in DO5 (arrow). (M–R) Stage 16 embryos stained for β3-tubulin (blue), βPS integrin (green), to visualize tendon cell-muscle connections and moeGFP (red) expressed under control of a DA3-specific col CRM (colLCRM), abbreviated col-gfp. Only βPS integrin and moeGFP are shown, except insets. (M) wt; DA3 ventral and dorsal attachment along the anterior and posterior segmental borders, respectively, are indicated by asterisks; inset, DT1 (arrowhead). (N) aop; DA3 with both DA3 and DA2-like (arrowhead) anterior attachments; DA3>DA2 transformation, inset. (O) edl: moeGFP expression in DA2 and DA3 (arrow and inset). The arrow indicates partial DA2>DA3 transformation; inset, bifid anterior DA3 attachment. (P) eya: moeGFP is lost in most segments or indicates partial or complete (inset) DA3>DA2 transformation. (Q) noc: DA3>DT1 transformation, resulting in DT1 (arrowhead in inset) duplication. (R) so: moeGFP expression in DO5; DO5>DA3 transformation (arrowhead) in some segments. (S–X) Schematic diagram of the most frequent DA2 and DL muscle phenotypes in aop, edl, eya, noc and so mutants; Col expression is in red; see Figure 2—source data 1 for statistics. Bars: 30 μm
-
Figure 2—source data 1
Quantification of muscle phenotypes observed in aop, edl, eya, noc and so mutant stage 15 embryos.
DA3>DA2: complete or partial DA3 into DA2 muscle orientation; DA2>DA3: complete or partial DA2 into DA3 muscle orientation; ColexpDO5: DO5 muscle expressing Col. Loss of Col: loss of DA3 muscle Col expression. DA3>DT1 transformations have been quantified in colLCRM-moeGFP embryos.
- https://doi.org/10.7554/eLife.14979.006
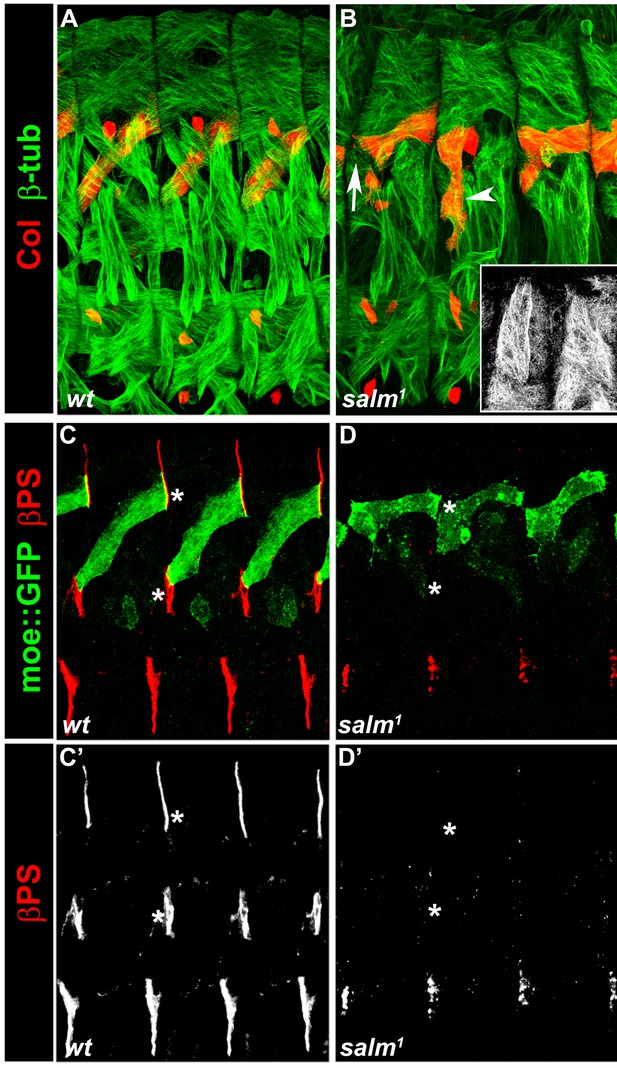
salm1 is required for proper skeletal attachment and morphology of the DA3 muscle.
(A,C,C’) wt, and (B,D,D’) salm1mutant stage 16 embryos. (A,B) Staining for Col (red) and β3-tubulin (green). (B) the white arrow and arrowhead point to a loose DA3 anterior attachment and a vertical DA3 fiber with no posterior attachment, respectively; the inset shows dorsal muscles without defined posterior attachment sites. (C,D) Staining for MoeGFP expressed under control of a DA3-specific CRM (colLCRM) (green) and βPS integrin (red). No βPS integrin accumulation is detected at the position of DL muscle attachment sites (white asterisks) in salm1 mutant embryos (C’,D’).

Snapshots for Videos 1–6.
Snapshots of Videos 1–6 , showing β3-tubulin staining of muscles (green) and Col expression (red) in stage 16 embryos. Five segments are shown in (A–D), two in (E–G). (A,E–E”) wt embryos, Video 1. (B) aop1 mutant, Video 2. (C) edll19, Video 3. (D) noc35ba2, Video 5. (F) eyaCII/Df(2L)BSC354, Video 4. (G) so3, Video 6. The positions of DA3, and DA2 and DA3 muscles are indicated in A,F,G, and C, respectively. DA2 is circles in A and LL1 in E,F. E,E”, F,F”, and G,G” are single channel in black and white, E’F’,G’ green chanel, E”,F”,G” red channel. DO5>DA3 and LL1>DA3 transformations are indicated in F,F” and G,G”, and F, respectively.
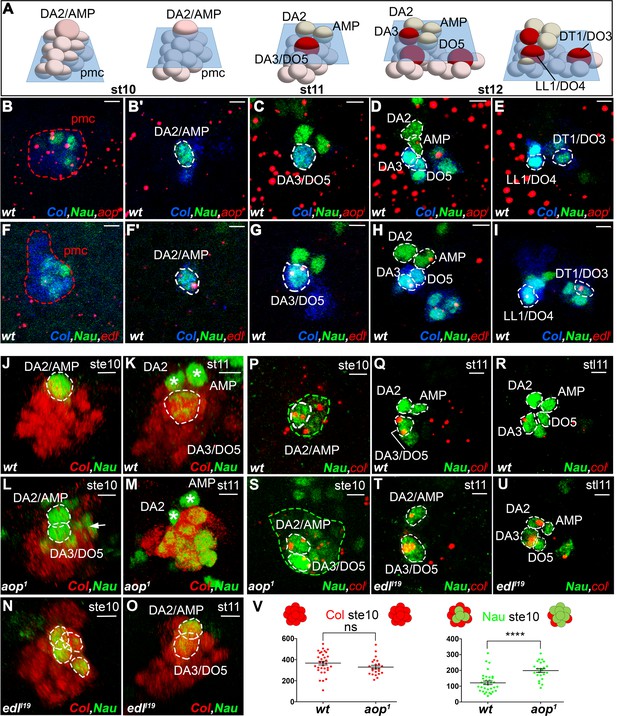
aop and edl differential expression and roles during PC selection.
(A) Schematic representation of the positions of DL PCs and FCs, relative to the A/P, D/V and proximal/distal axes in stage 10, 11 and 12 wt embryos (Video 7); the blue trapeziums indicate planes of section shown in panels (B–K) and (R–U); Col expression is in red. (B–I) ISH to aop (B–E) and edl (F–I) primary transcripts (red dots), in wt embryos stained for Col (blue) and Nau (green), at stages indicated above; early stage is abbreviated ste; two different planes of the same embryo are shown in B and B’, F and F’. aop transcription in the Col PMC (B), and the AMP (D). (F–I) edl transcription in all PCs, the AMP and the DA3 FC. (B,F) Nau accumulation in two to three Col PMC cells, below the emerging PC. (J,O) 3D reconstruction of the Col PMC during DA2/AMP and DA3/DO5 PC selection; Col staining, red, Nau, green. The embryonic stage indicated in each panel. (J, K) wt; (J) apical DA2/AMP PC (dotted white circle); (K) apical DA3/DO5 PC (dotted circle), DA2 FC and AMP (asterisks). (L, M) aop embryos; (L) premature DA3/DO5 PC selection; additional Nau-expressing PMC cells (arrow), become PCs, (M). (N, O) edl embryos; (N) no PC is selected; a group of 3 to 4 Nau-expressing cells is embedded in the Col PMC; (O) two PCs are simultaneously selected. (P–U) ISH to col primary transcripts (red), Nau staining (green); (P–R) wt; sequential col transcription in the PMC and DA2/AMP PC (P), DA3/DO5 PC (Q), and DA3 FC (R). (S) aop mutant: simultaneous col transcription in two apical PCs; increased number of low level Nau-expressing cells (green dotted circle). (T,U) edl mutant; ectopic col transcription in the DA2/AMP PC (T) and DA2 FC (U). (V) Measurement of the diameter of Col (red) and Nau (green) expressing domains in early stage 10 wt and aop embryos. The Col expression domain is identical (P value = 0,1410; ns) and Nau domain expanded in aop compared to wt (P value<0,0001; ****), schematized on top of the statistics. Bars: 5 μm
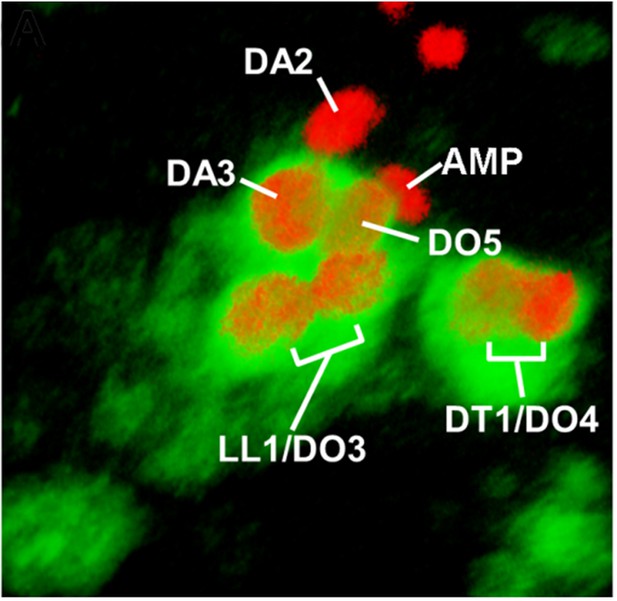
Snapshot for Video 7.
Stage 12 embryo. Col (green) is expressed in a large promuscular cluster and the FCs at the origin of the DA3, DO5, LL1, DO3, DT1 and DO4 muscles, indicated on the screenshot. Col expression has been lost from the DA2 FC and the AMP at this stage. All FCs and the AMP express Nau (red). The Antero-Posterior, Dorso-Ventral and Distal-Proximal axes are indicated on the video by red, green and blue arrows, respectively.

Transient Nau expression in subsets of PMC cells.
(A) Schematic representation of the positions of the DA2/AMP PC, relative to the A/P, D/V and proximal/distal axis in stage early 10 (st e10) and late 10 (st l10); the trapezium indicates planes of sections shown in panels (B–C). Nau expression is in blue. (B–C) wt embryos at st e10 (B) and st l10 (C) stained for Nau (blue) and dpERK (green). B and B' are two planes of section across a small equivalence group of cells both expressing Nau and accumulating dpERK. The selected PC (B') is positioned apical to the equivalence group (B). (C) At a slightly later stage, only the PC maintains Nau and dpERK (arrow).

Extended analysis of the aop muscle mutant phenotype.
(A, B) stage 16 embryos stained for Col (red) and F-actin (green). (A) wt, (B) aop1mutant embryo displaying several DA3 (horizontal arrow) or LL1 (vertical arrow) muscles.
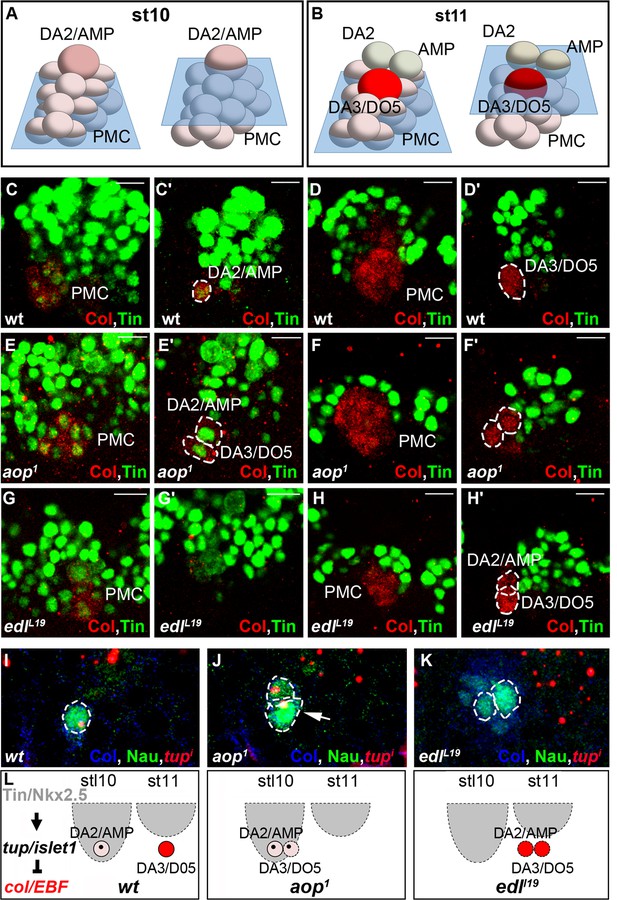
aop and edl control the temporal sequence of PC selection.
(A,B) Schematic representation of the DA2/AMP PC and FCs and DA3/DO5 PC, at stages 10 (A) and 11 (B); the blue trapeziums indicate the planes of section shown below. (C–H’) Tin (green) and Col (red) embryo staining. (C, C’) Tin expression in the DA2/AMP PC and underlying PMC cells. (D, D’) Tin expression has regressed dorsally; the DA3/DO5 PC and underlying PMC cells are Tin negative. (E–F’) aop mutants; (E, E’), stage 10, two Col and Tin-expressing PCs are selected; (F, F’) stage 11, Col positive, Tin-negative cells are selected. (G–H’) edl mutants; (G, G’), No PC is selected. (H, H’) Two PCs are selected after dorsal regression of Tin expression. (I–K) FISH to tup primary transcripts in wt (I) aop (J) and edl (K) stage 10 (I, J) and 11 (K) embryos, stained for Col (blue) and Nau (green). In wt embryos (I) tup expression is only detected in the first selected Col positive PC (DA2/AMP, 100% n = 27). In aop mutants (J), tup transcription is sometimes detected (17% n = 34) in a second Col positive PC (arrow). In edl mutants (K), tup transcription is frequently lost in Col positive PCs (82% n = 29). (L) Summary scheme of the aop and edl phenotypes; wt, late stage (stl) 10; only the first-born, DA2/AMP PC inherits Tin, and activates tup, preventing Col autoregulation (left) which occurs in the second born, DA3/DO5 PC, in absence of Tin and Tup, stage 11 (Boukhatmi et al., 2012). In edl and aop mutants, the temporal sequence of PC selection is compromised; it occurs too early and too late in aop and edl mutants, respectively, leading to confusions of DA2 and DA3 fates. Bars: 10 μm
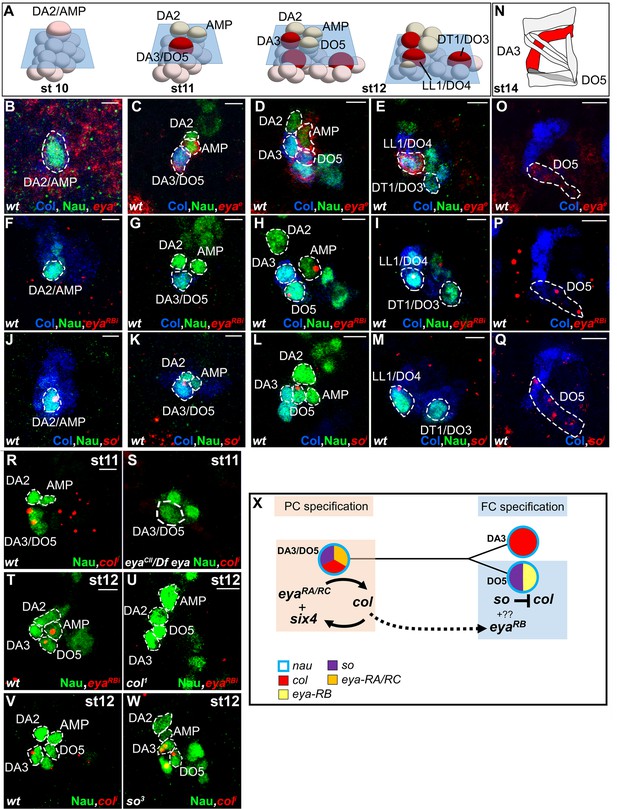
Sequential eya and so transcription and control of col transcription in distinct muscle lineages.
(A) Schematic representation of the positions of DL PCs and FCs in stage 10, 11 and 12 wt embryos, reproduced from Figure 3A; the blue trapeziums indicate the planes of section shown below, panels (B–M). (B–M and O–Q) ISH to eya and so transcripts (red) in wt embryos stained for Col (blue) and Nau (green). (B–E), eya expression in the DA2/AMP (B), DA3/DO5 (C) AMP and DO5 FC (D) LL1/DO3 PC (E). (N) Schematic representation of the DL muscle pattern, DA3 in red and DO5 in grey. (O) DO5 eya expression. (F–I) eya-RB transcription in the AMP DO5 FC (H) and LL1/DO4 PC (I). (P) DO5 eya-RB transcription. (J–M, Q) so transcription in the DA2/AMP, DA3/DO5 and LL1/DO4 PCs, DO5 FC and muscle. (R,S) Loss of col transcription in the DA3/DO5 PC in eya mutants. (T–U) Loss of eya-RB transcription in col mutants (U). (V,W) col ectopic transcription in the DO5 FC in so mutants. Embryos in R-W are co-stained for Nau (green). (X) Summary diagram of eya and so expression and function in DL muscle lineages. Bars: 5 μm
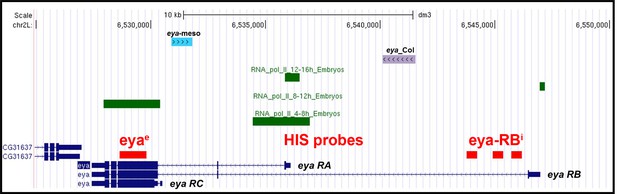
Schematic representation of the eya genomic region and transcripts.
Schematic representation of the eya genomic region chromosomal positions Chr2L:6,525,000–6,550,000. The different eya transcripts (RA, RB, RC) initiated from different Transcription Start Sites are indicated. Red boxes indicate the position of the exonic (eyae) and intronic (eya-RBi) probes used for FISH experiments. Green boxes show RNA-PolII binding at the indicated embryonic stage (Zinzen et al., 2009). Two previously characterized mesodermal CRMs are indicated by blue (Liu et al., 2009) and purple boxes, respectively. eya_Col CRM activity depends upon in vivo Col binding (de Taffin et al., 2015).
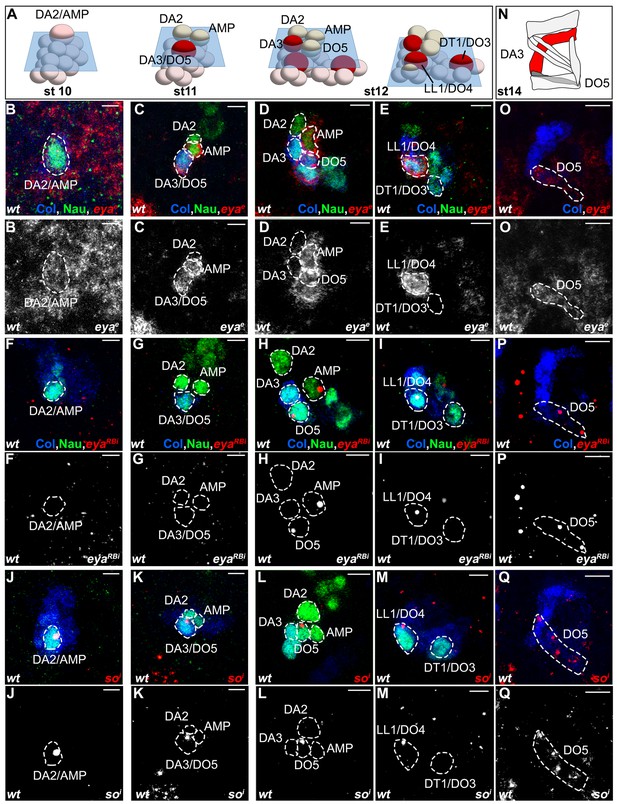
Sequential eya and so transcription.
(A) Schematic representation of the DL PCs, FCs and muscles as in Figure 5. (B–Q) ISH to eya and so transcripts (red) in wt embryos stained for Col (blue) and Nau (green). To complement Figure 5, the red channel is show separately in black and white.
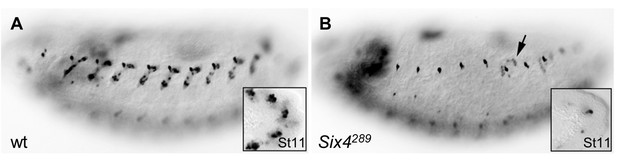
Loss of DA3 Col expression in Six4 mutant embryos.
(A) Stage 15 wt, and (B) homozygous mutant embryos for a null allele of Six4, Six4289. Col expression is detected in 100% of segments in wt embryos (n = 117), and lost in 84% of segments in Six4 mutants (n = 249). In most of the remaining muscles, residual Col expression indicates a DA3>DA2 transformation (arrow in B). Col expression is already lost at the PC stage in Six4 mutants (insets in A,B), similar to the loss observed in eya mutants (See Figure 2D).
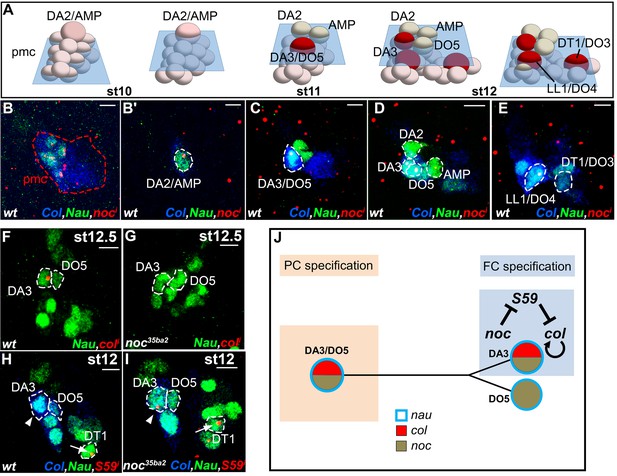
noc transcription and control of col and S59 transcription in DL muscle lineages.
(A) Relative positions of DL PCs and FCs between stages 10 and 12, reproduced from Figure 3A; the blue trapeziums indicate the planes of section shown below, panels (B–E). (B–E), Embryos co-stained for Nau (green) and Col (blue); (B) noc transcription (red) in a small subset of Col PMC cells expressing low Nau level (B), the DA2/AMP (B’), DA3/DO5 and DA3 and DO5 FCs (C,D) and LL1/DO4 but not the DT1/DO3 PC (E). (F–I) Embryos stained for Nau (green) and (H,I) Col (blue). (F, G) loss of col transcription in the DA3 FC (red dots) in noc mutants. (H) wt S59 transcription in DT1 (red dots, arrow) and (I) ectopic transcription in the DA3 FC (arrowhead) in noc mutant embryos. (J) Summary diagram of noc expression and function in DL muscle lineages. Bars: 5 μm

S59 represses col transcription.
(A, B) stage 16 embryos stained for Col (red) and β3-tubulin (green). (A) wt, (B) embryo expressing S59 under control of the twist-gal4 mesodermal driver. The muscle pattern is strongly disorganized, and Col muscle expression is lost. (C,D) stage 13 embryos stained for col primary transcripts (red), Col (green) and Nau (blue). (C) wt; the arrow indicates the posterior group of cells giving rise to the DT1 and DO3 FCs. At this stage, col expression is rarely detected in these cells (6% of segments, n = 52). (D) S59 mutant embryo. col expression is maintained in at least one posterior cell, likely the DT1 FC (81% of segments, n = 65).
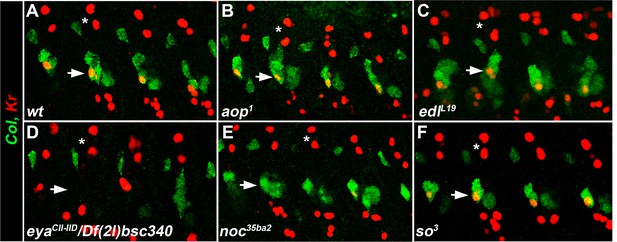
eya and noc are required for Kr expression in the LL1 FC.
(A–F) Stage 12 embryos stained for Col (green) and Kr (red). (A) wt, co-expression of Col and Kr in the LL1 FC (horizontal arrow). Kr expression is detected in aop1 (B), edlL19 (C) and so3 (F), and lost in eyaCII/Df(2L)BSC354 (D) and noc35ba2 (E) mutant embryos. Of note, Kr expression is lost in DL, but not dorsal FCs (asterisks).
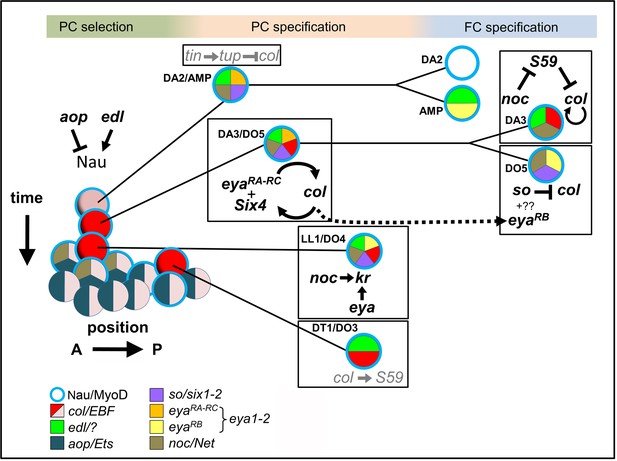
Intertwined transcriptional control of DL muscle identity: A progressive resolution of the possible.
Diagrammatic representation of transcription regulatory interactions and loops operating in DL muscle identity specification. One abdominal segment is considered. 3 steps are indicated on top and color-shaded. Left, PC selection; the A/P axis is on the abscissa and the developmental time on the ordinate. aop and edl positively and negative regulate Nau expression (blue circle) in a subset of PMC cells, favoring and inhibiting selection of PCs from PMC cells, respectively. Center, PC specification: the dorsal DA2/AMP and dorso-lateral DA3/DO5, LL1/DO4 and DT1/DO3 PCs are represented. Right, FC specification; only the DA3 and DO5 lineages are detailed. Color coding of expression of the different iTFs, and the names of their vertebrate orthologs are indicated. Genes and either positive (arrow), or negative (crossed line) regulatory steps identified in this study are drawn in black; previously reported interactions are in grey.





