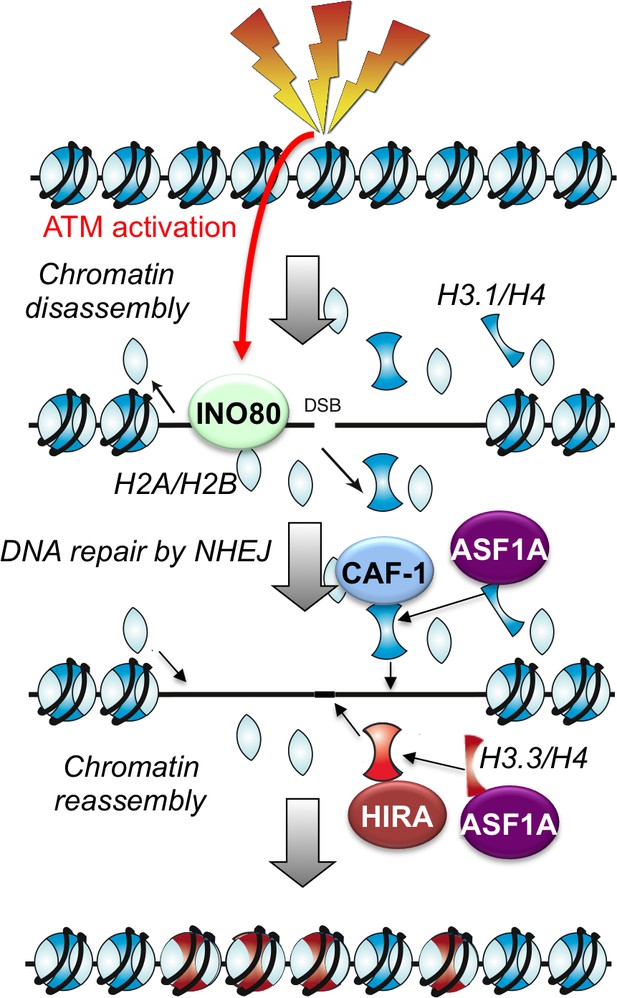
Nucleosome disassembly during human non-homologous end joining followed by concerted HIRA- and CAF-1-dependent reassembly
Figures
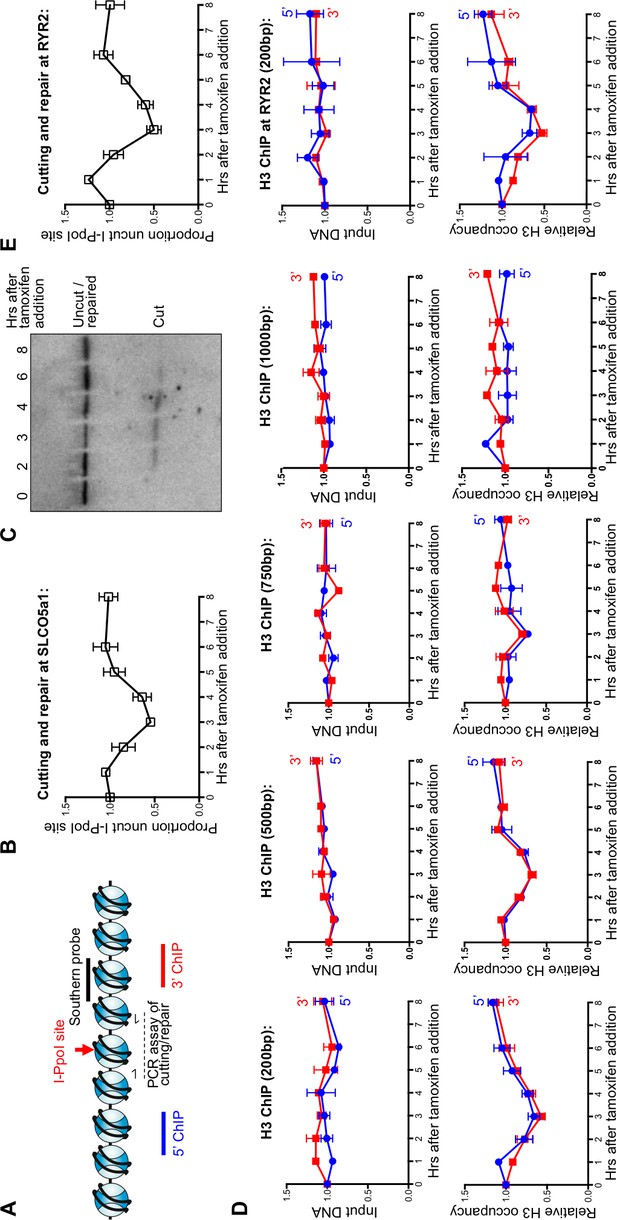
Local nucleosome disassembly and reassembly around DSBs in human cells.
(A) Schematic demonstrating primer pairs spanning the I-PpoI site that were used to measure cutting and repair by quantitative PCR, while ChIP analysis was performed on the chromatin to the 5’ and 3’ of the I-PpoI site. (B) The kinetics of generation and repair of the DSB induced in the SLCO5A1 gene by the I-PpoI endonuclease. Shown is the average +/- SEM from three independent experiments. (C) Southern analysis of the kinetics of generation and repair of the DSB induced in the SLCO5A1 gene by the I-PpoI endonuclease from an independent time course from that shown in (B). (D) Quantitation of proportion of input DNA and H3 occupancy from ChIP analysis at 200, 500, 750 and 1000 bps (from left to right panels) away from the I-PpoI site within the SLCO5A1 gene. Shown is the average +/- SEM from three independent experiments. (E) (from top to bottom) DNA cutting analysis at the I-PpoI site within the RYR2 gene, input and H3 occupancy from H3 ChIP analysis 200bp from the I-PpoI site within the RYR2 gene. Shown is the average +/- SEM from three independent experiments.
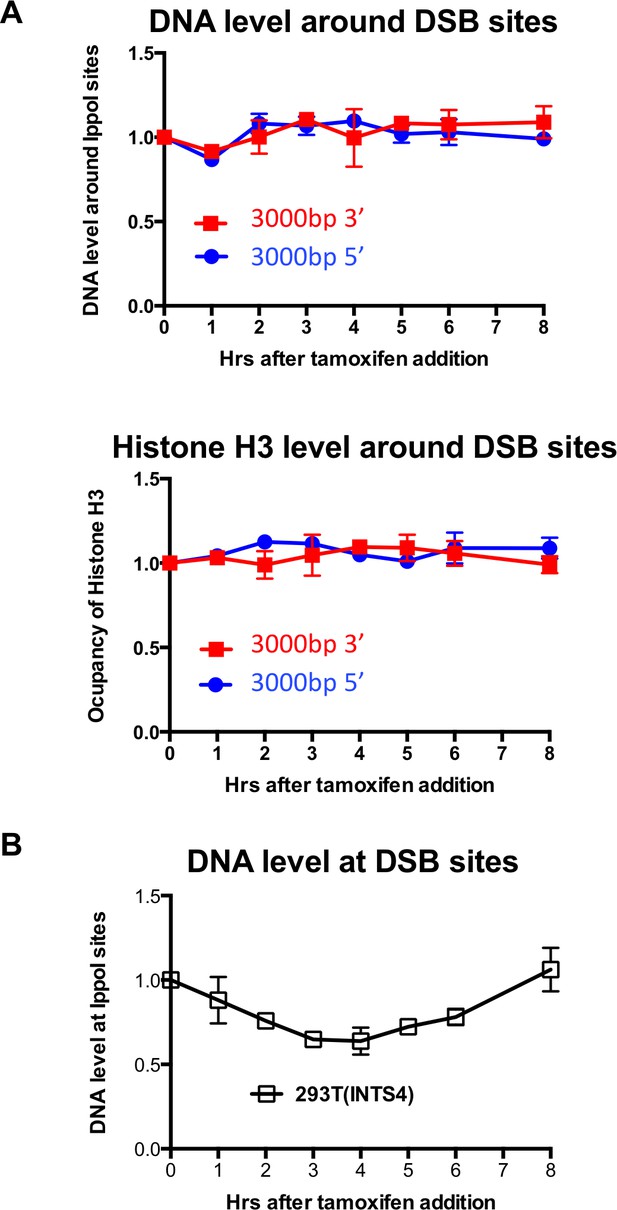
Limited H3 disassembly at a distance and I-PpoI cleavage at INTS4.
(A) Input and histone H3 occupancy from the H3 ChIP assayed at 3000 bp from the I-PpoI site at SLCO5A1. This is from the same experiment shown in Figure 1, but using primer pairs at a greater distance. (B) Quantitation of cutting and repair analysis at the INTS4 gene.
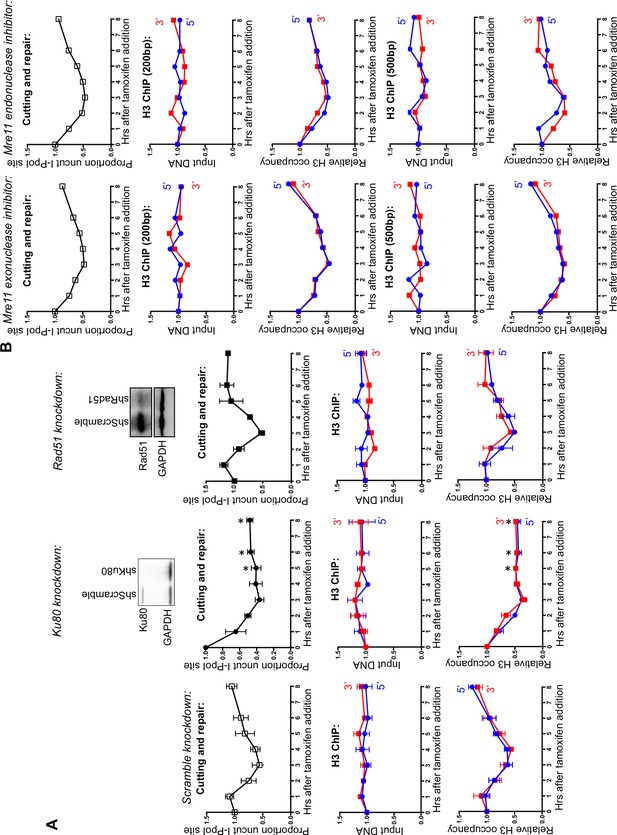
DNA resection is not required for histone H3 disassembly.
(A) The repair of the I-PpoI sites is by NHEJ not HR. At the top is shown western-blotting of the RAD51 and KU80 knockdowns; below is shown the kinetics of generation and repair of the DSBs at SLCO5A1 I-PpoI sites in scramble shRNA knockdown (left), sh-hKU80 (center) and sh-hRAD51 (right) knockdown cells respectively. At the bottom is shown quantitation of the proportion of input DNA and H3 occupancy from ChIP analysis at 200 bps from the I-PpoI site within the SLCO5A1 gene in the scrambled shRNA knockdown (left panels), the KU80 knockdown (center panels) and RAD51 knockdown (right panels) knockdown cells. Shown is the average +/- SEM from three independent experiments. Asterisks indicate significant changes from the scramble shRNA knockdown, p = 0.005, as determined by the Student’s t-test. (B) Inhibition of MRE11 has no effect on repair or chromatin disassembly / reassembly around the I-PpoI site. The panels on the left are experiments performed with cells grown with the MRE11 exonuclease inhibitor PFM39, while those on the right were from experiments performed with cells grown with the MRE11 endonuclease inhibitor PFM03. At the top is shown the kinetics of generation and repair of the DBSs at SLCO5A1 I-PpoI sites; below is the quantitation of proportion of input DNA and H3 occupancy from ChIP analysis at 200 and 500 bps from the I-PpoI site within the SLCO5A1 gene.
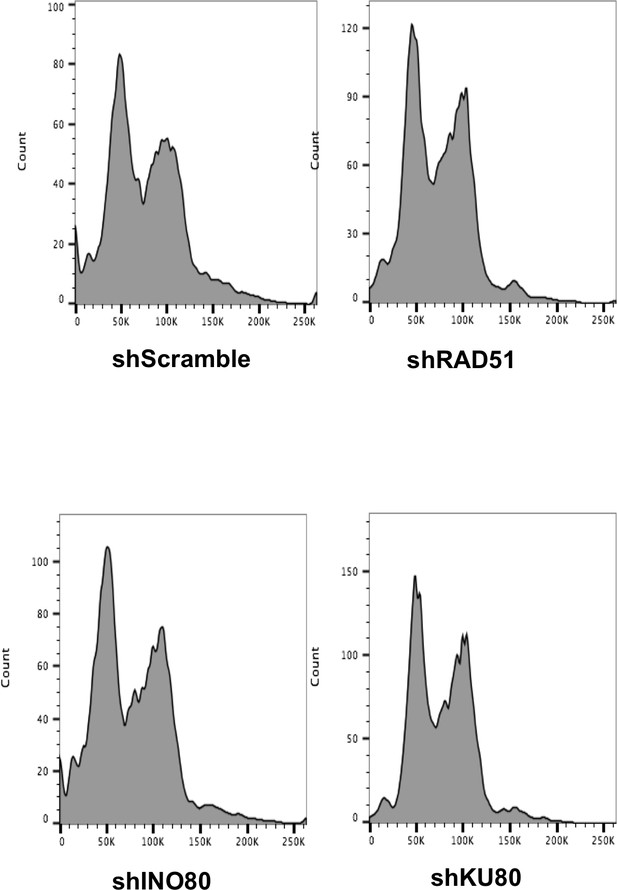
Flow cytometry analysis of propidium iodide stained DNA, in cells with the indicated knockdowns.
https://doi.org/10.7554/eLife.15129.006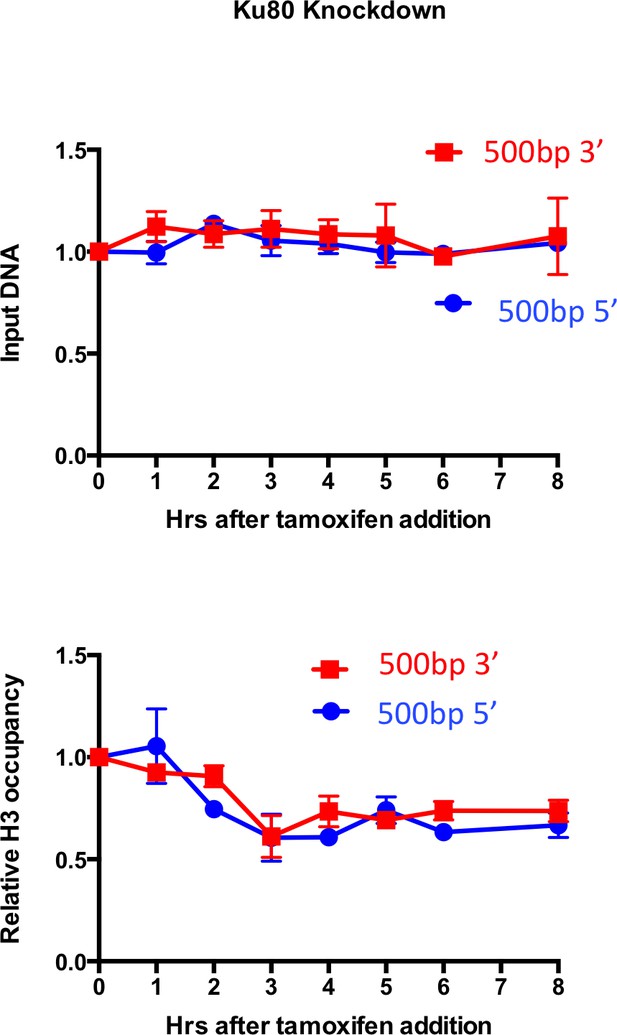
H3 disassembly in the absence of KU80.
(A) Input and histone H3 occupancy from the H3 ChIP assayed in KU80 knockdown cells at 500bp from the I-PpoI site at SLCO5A1. This is from the same experiment shown in Figure 2, but using primer pairs at a greater distance.
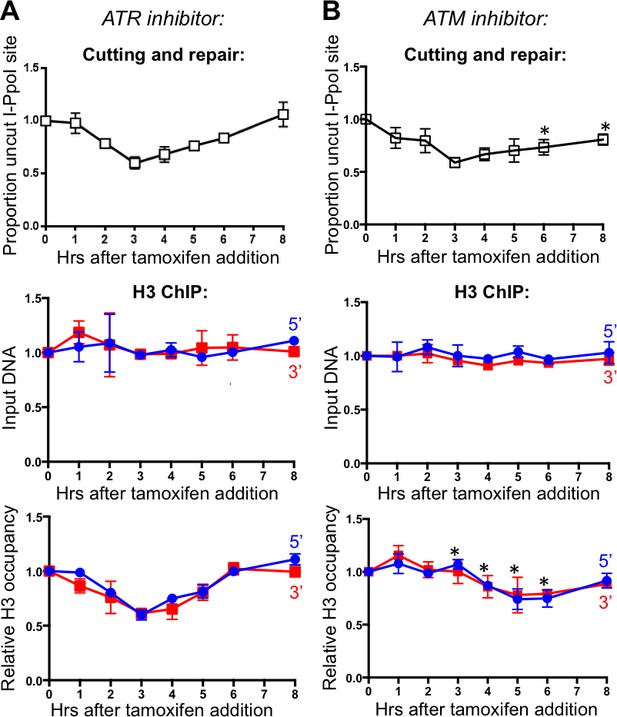
ATM, but not ATR, promotes histone H3 disassembly.
(A) The kinetics of generation and repair of the DBSs at SLCO5A1 I-PpoI sites is shown at the top; and quantitation of proportion of input DNA and H3 occupancy from ChIP analysis at 200 bps from the I-PpoI site within the SLCO5A1 gene in cells treated with ATR inhibitor VE-821 is shown below. Shown is the average +/- SEM from three independent experiments. (B) As in A, but with treatment with ATR inhibitor KU55933. Asterisks indicate significant changes from no inhibitor, p=0.005, as determined by the Student’s t-test.
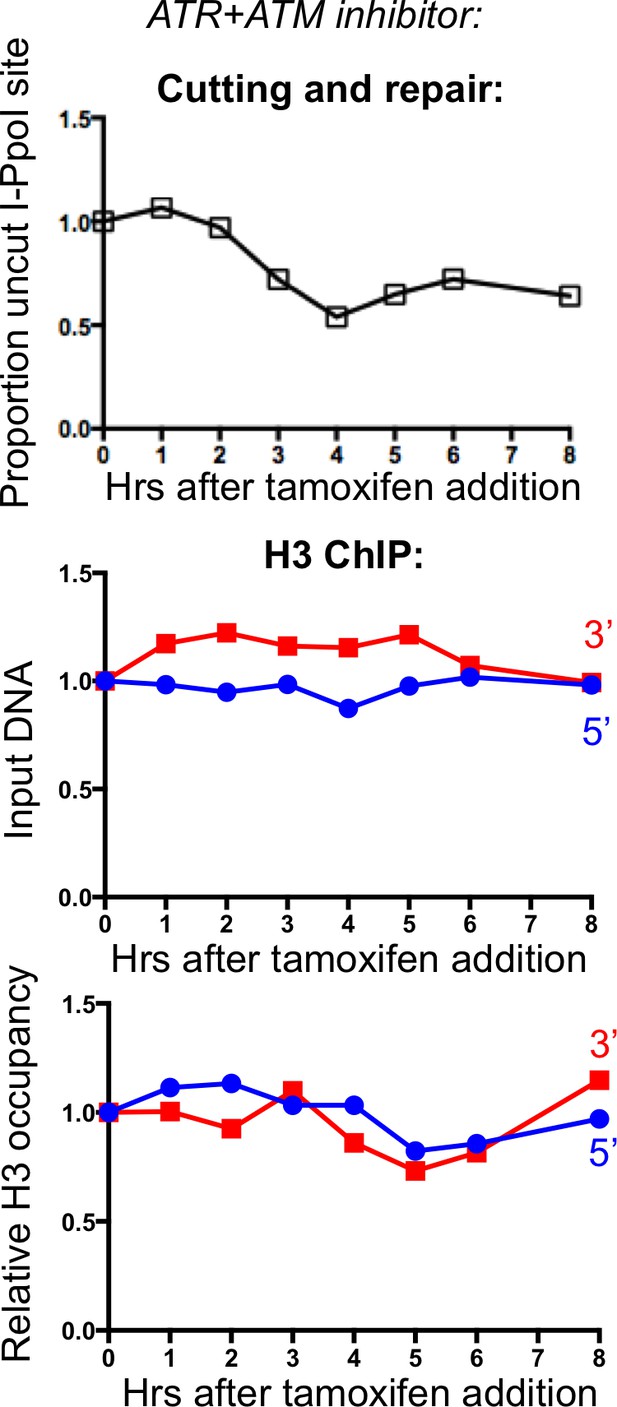
Effect of combined ATM and ATR inhibitors on chromatin disassembly.
The same inhibitors shown in Figure 3 were used in combination, and no additional affect beyond ATM inhibition alone was noted.
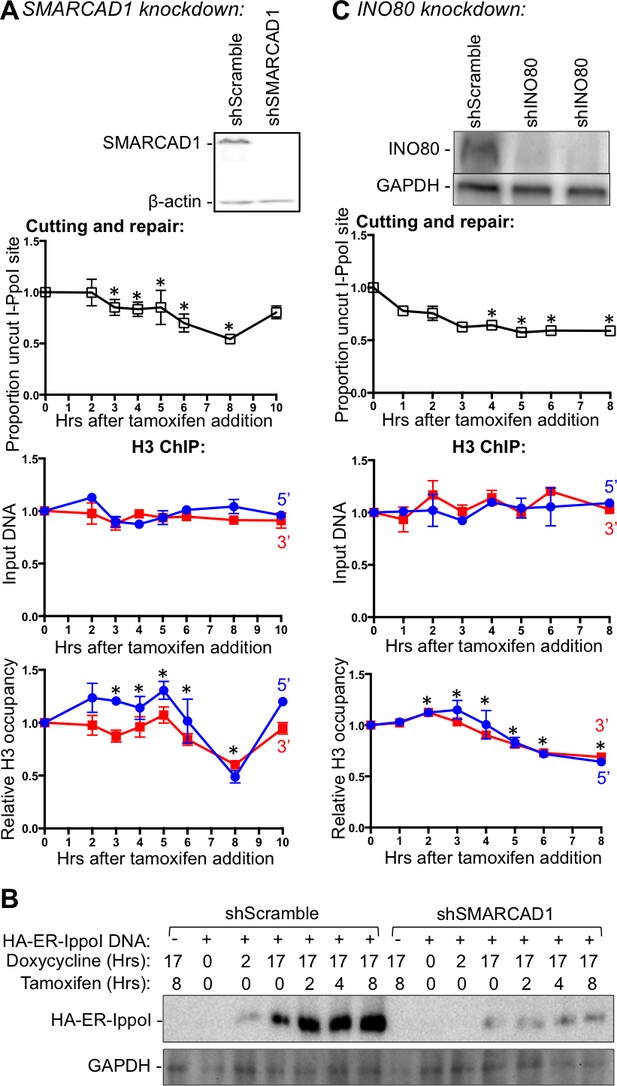
INO80, but not SMARCAD1, regulates histone H3 disassembly.
(A) Western-blotting of SMARCAD1 knockdown is shown at the top. The kinetics of generation and repair of the DBSs at SLCO5A1 I-PpoI sites is shown below; and quantitation of proportion of input DNA and H3 occupancy from ChIP analysis at 200 bps from the I-PpoI site within the SLCO5A1 gene in SMRACAD1 knockdown cells is shown at the bottom. Shown is the average +/- SEM from three independent experiments. Efficient cutting and repair was seen in control knockdown cells performed in parallel (data not shown). Asterisks indicate significant changes from the scramble shRNA knockdown, p = 0.005, as determined by the Student’s t-test. (B) Kinetics of I-PpoI expression in Scrambled shRNA treated and SMARCAD1 knockdown cells respectively. (C) As in A, but for INO80 knockdown.
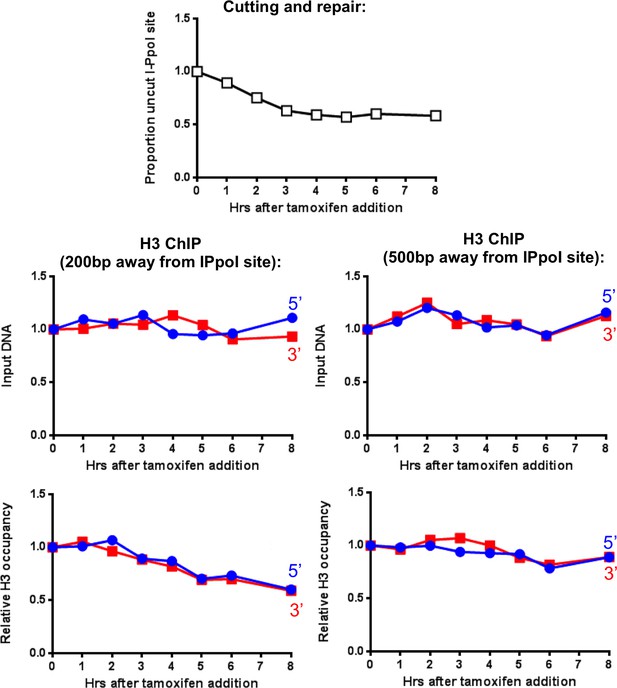
Effect of combined ATM inhibition and INO80 knockdown on chromatin disassembly is no more severe that INO80 knockdown alone.
https://doi.org/10.7554/eLife.15129.011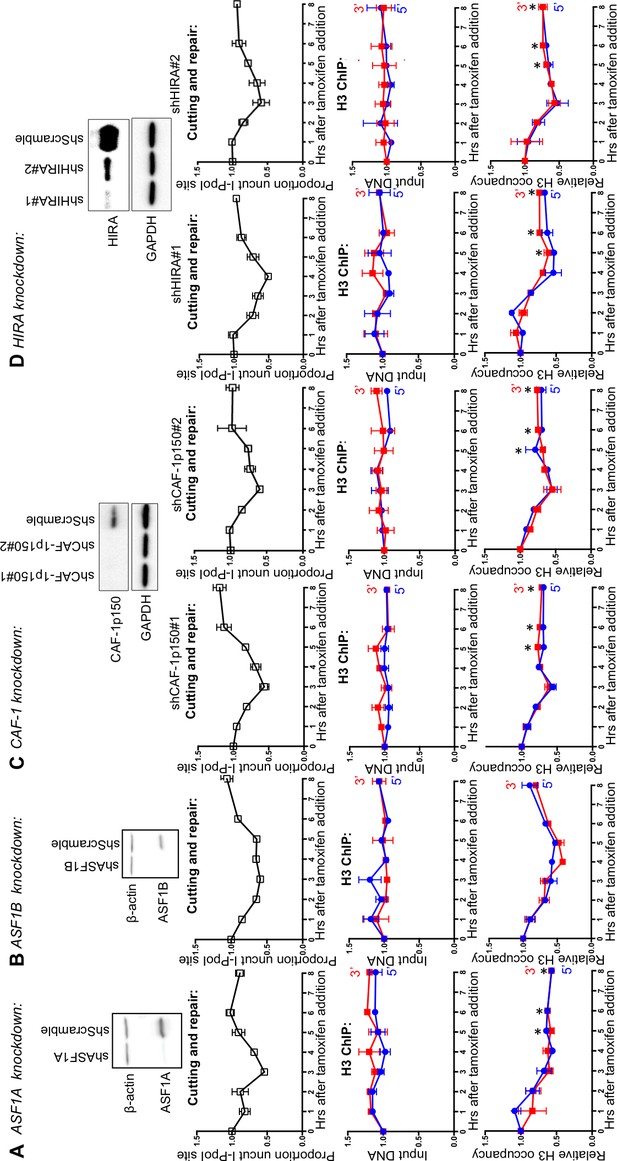
ASF1A, CAF-1 and HIRA promote chromatin reassembly.
(A) Western-blotting of ASF1A knockdown is shown at the top. The kinetics of generation and repair of the DBSs at the SLCO5A1 I-PpoI site is shown below; and quantitation of proportion of input DNA and H3 occupancy from ChIP analysis 200 bps from the I-PpoI site within the SLCO5A1 gene in ASF1A knockdown cells is shown at the bottom. Shown is the average +/- SEM from three independent experiments. Asterisks indicate significant changes from the scramble shRNA knockdown, p=0.005, as determined by the Student’s t-test. (B) As in A, but for ASF1B. (C) As in A, but for CAF-1. Data on the left and right are from two independent shRNAs as indicated. (D) As in C, but for HIRA.