Diagnostic potential of tumor DNA from ovarian cyst fluid
Figures
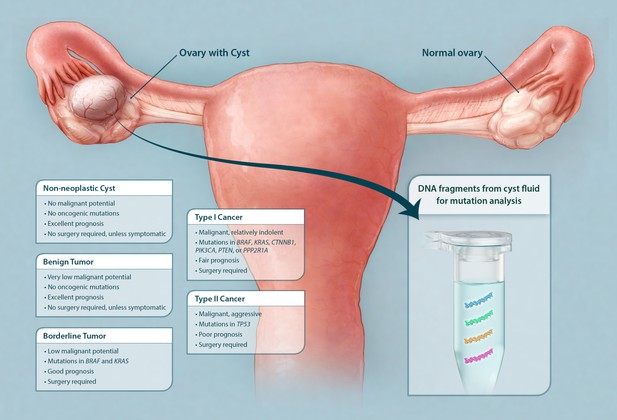
Schematic showing classes of ovarian cysts and the diagnostic potential of the cyst fluid.
Ovarian cysts and tumors are currently classified according to microscopic evaluation after surgical removal. The majority of ovarian cysts are non-neoplastic (often 'functional' in premenopausal women). Ovarian tumors with combined cystic and solid components are either benign tumors, borderline tumors, or malignant cancers (type I or II). Only cysts associated with borderline tumors and cancers require surgical excision. We show here that the DNA purified from cyst fluid can be analyzed for somatic mutations commonly found in their associated tumors. The type of mutation detected not only could indicate the type of tumor present but also could potentially inform management.
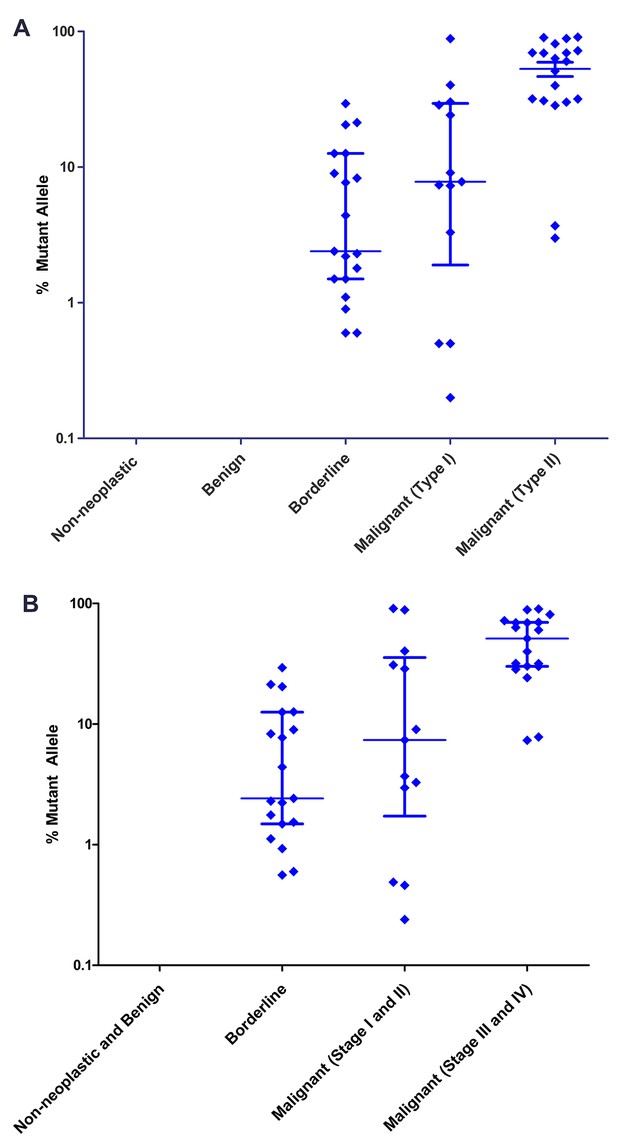
Mutant allele fractions.
(A) Classification by tumor type. No mutations were found in the DNA of non-neoplastic or benign cysts. Of the cysts that required surgery, the median mutant allele fraction was higher in the cyst fluids associated with type II cancer (60.3%) than type I (7.8%) or borderline tumors (2.4%). (B) Classification by tumor stage. The DNA from cyst fluids of late-stage cancers had a higher median mutant allele fraction (51.2%) than those of early-stage cancers (7.4%) or borderline tumors (2.4%). Percent mutant allele is depicted on a logarithmic scale. Horizontal bars depict median and IQR.
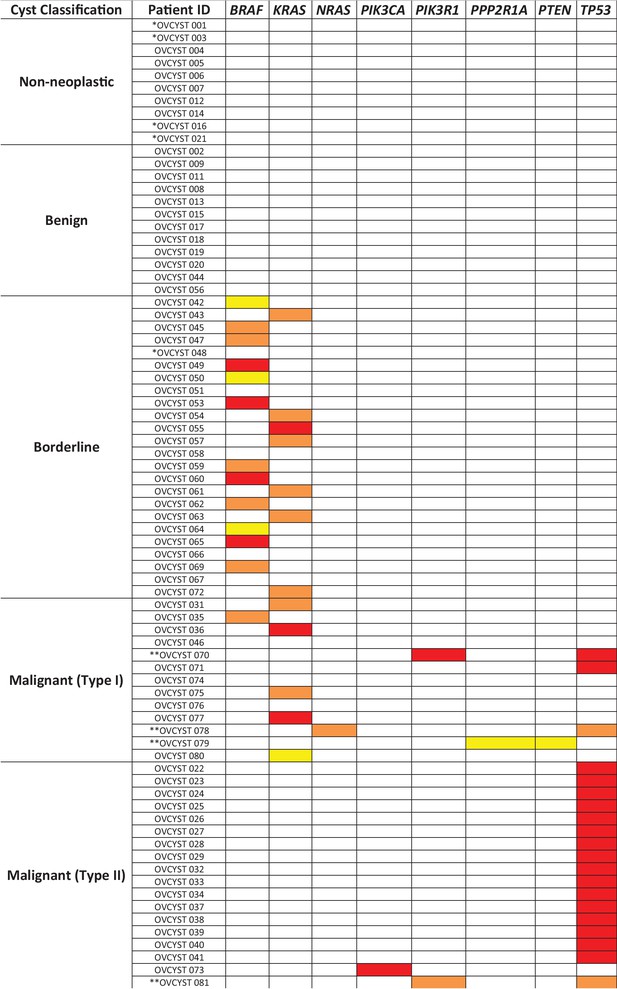
Mutated genes found in the cyst fluid samples.
Yellow boxes represent mutations with mutant allele frequency (MAF) between 0.1% and 1%; orange boxes represent mutations with MAF between 1 and 10%; red boxes represent mutations with MAF greater than 10% (* indicates patients with insufficient DNA for analysis; ** indicates patients with two detected mutations).
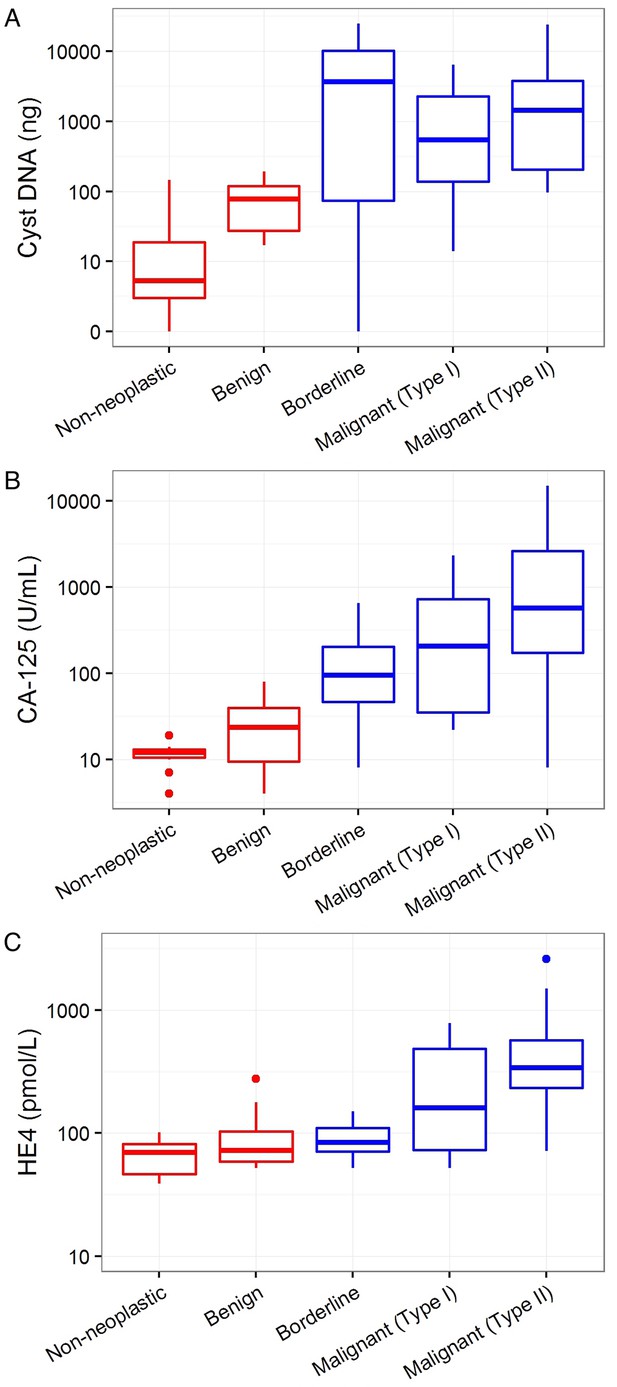
Markers associated with the need for surgery.
Cyst DNA amount and levels of commonly used ovarian cancer serum biomarkers are plotted according to the cyst type and need for surgery. (A) The amounts of DNA in cyst fluids was generally higher in cysts requiring surgery (blue) than those that do not (red), but no significant correlation was found (p=0.69). (B) CA-125 levels was significantly higher in cysts that required surgery than those that do not (p=0.01). (C) Serum HE4 levels was not correlated with the need for surgery (p=0.92). All values are depicted on a logarithmic scale. P-values were calculated using Firth’s penalized likelihood logistic regression in a multivariate model (See Materials and methods).
Tables
Detection of tumor-specific mutations in cyst fluid. The fraction of samples detected and the median fraction of mutant alleles are indicated, grouped by cyst type, cancer stage, and the need for surgery.
| Fraction of samples detected (95% confidence interval) | Median fraction of mutant alleles (IQR) | Total # of samples | |
|---|---|---|---|
| Type | |||
| Non-neoplastic | 0% (0–46%) | 0% (0–0%) | 6 |
| Benign tumor | 0% (0–26%) | 0% (0–0%) | 12 |
| Borderline tumor | 83% (61–95%) | 2.4% (1.5–10.8%) | 23 |
| Type I cancer | 77% (46–95%) | 7.8% (3.3–28.7%) | 13 |
| Type II cancer | 100% (81–100%) | 60.3% (31.3–70.8%) | 18 |
| Cancer stage | |||
| Early (I and II) | 82% (48–97%) | 7.4% (3.0–30.9%) | 11 |
| Late (III and IV) | 95% (75–100%) | 51.2% (30.2–69.5%) | 20 |
| Cysts requiring surgery | |||
| No | 0% (0–26%) | 0% (0–0%) | 18 |
| Yes | 87% (75–95%) | 12.6% (2.7–40.2%) | 54 |
Multivariate analysis for markers associated with need for surgery. The presence of a mutation, cyst DNA amount, and common serum biomarkers for ovarian cancer were analyzed for association with cysts that require surgical removal (Firth’s penalized likelihood logistic regression).
| Criteria | p value |
|---|---|
| Mutation present | <0.001 |
| Serum CA-125 elevated | 0.01 |
| HE4 elevated | 0.92 |
| Cyst DNA amount | 0.69 |
Additional files
-
Supplementary file 1
Patient demographics.
- https://doi.org/10.7554/eLife.15175.009
-
Supplementary file 2
Mutations identified in cyst fluids and associated tumors.
- https://doi.org/10.7554/eLife.15175.010
-
Supplementary file 3
Primer sequences used in multiplex assay.
- https://doi.org/10.7554/eLife.15175.011





