Cooperative unfolding of distinctive mechanoreceptor domains transduces force into signals
Figures
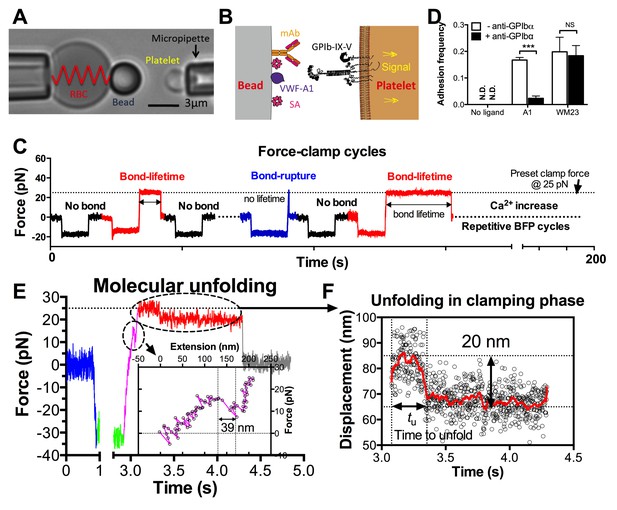
BFP analysis of ligand binding kinetics and domain unfolding mechanics of platelet GPIbα.
(A) BFP micrograph. A micropipette-aspirated RBC with a probe bead attached to the apex (left) was aligned against a platelet held by an opposing micropipette (right). (B) BFP functionalization. The probe bead was coated with streptavidin (SA, for attachment to the biotinylated RBC) and VWF-A1 or mAb (left) for interaction with platelet GPIb (right). (C) Representative force vs. time traces of repetitive force-clamp cycles over a 200-s period. Cycles produced different results are color-coded (black: no bond; blue: bond-rupture; red: bond-lifetime). (D) Mean ± s.e.m. of adhesion frequencies (n ≥ 3) of platelets binding to beads functionalized with indicated proteins in the absence (open) or presence (closed) of 50 µg/ml AK2. The coating densities are 131 and 95 µm-2 for A1 and WM23 respectively. *** = p < 0.001 by Student t-test. (E) Force vs. time trace of a representative BFP cycle showing unfolding signatures in both ramping and clamping phases. The inset zooms in the ramped unfolding signature and indicates the unfolding length. (F) Zoom-in view of the clamped unfolding signature in (E). Higher displacement resolutions were obtained after smoothing the raw data (points) by the Savitzky-Golay method (curves). Time to unfolding (tu) is indicated.
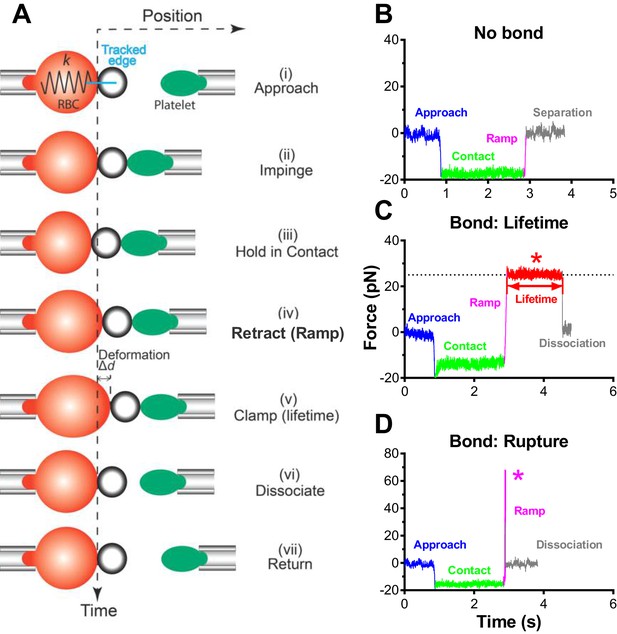
BFP test cycle.
(A) Schematic of a force-clamp experiment mode. Position (horizontal axis) of the probe bead attached to the apex of the RBC aspirated by the fixed micropipette (left) goes through indicated stages (i)-(vii) of a force-clamp test cycle as time (vertical axis) increases in response to the interaction with the target platelet aspirated by the moving micropipette (right) driven by a piezo actuator and controlled by a computer. The origin of the horizontal axis indicates the resting position of the RBC apex when no force is applied to it. Upon contact, the platelet touches the probe bead and changes its position (<0 by impinging (compressive force) and >0 by pulling (tensile force)), causing RBC deflection that can be modeled by a spring with a spring constant k to allow conversion of the deformation (∆d) to force (= k × ∆d). The probe bead position is tracked with high temporal resolution using a high-speed camera equipped with real-time image analysis. The line edge tracker of the RBC apex position is shown in blue. (B–D) Representative force vs. time traces illustrating three different outcomes of a BFP test cycle. Different segments corresponding to different stages are indicated using matched colors. A platelet was driven to approach (blue), contact and impinge (green) a probe bead, and retract (magenta). When no bond resulted, the platelet separated from the bead (gray) and returned to the original position (B). When a bond resulted and survived the tensile ramped force (magenta) to a preset level (in the force-clamp mode), the computer program stopped retracting the platelet and held its position to a clamp force (marked by *) for bond lifetime measurement (red) until dissociation (gray) after which the platelet returned to the original position (C). When a bond resulted but did not survive the ramping phase (magenta), the bond was ruptured (marked by *) after which the platelet returned to its original position (gray) (D).
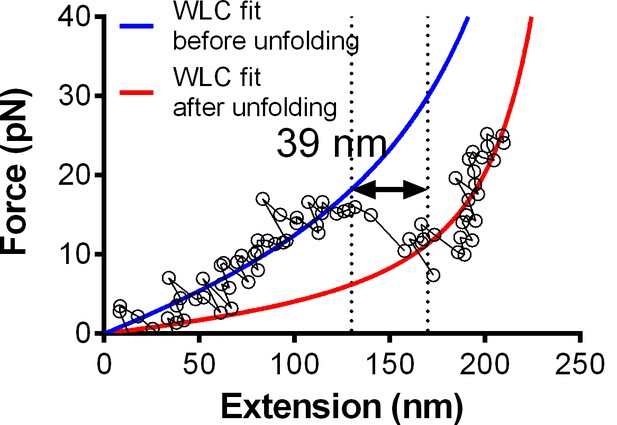
Fitting of the WLC model to the force-extension traces (Figure 1E insert) before (blue) and after (red) the observed GPIbα ramped unfolding event.
The unfolding length can be estimated directly (indicated) or calculated from the difference between the two best-fit contour lengths (318.6–279.3 = 39.3 nm). Both give comparable results.
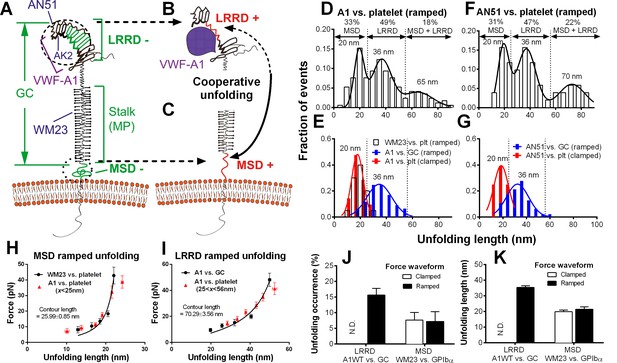
Identification and characterization of unfolding of LRRD and MSD.
(A–C) Schematics of GPIbα on the platelet membrane (A), highlighting the folded (−) and unfolded (+) LRRD (B) and MSD (C). Different regions and binding sites for VWF-A1 and mAbs are indicated. (D–G) Normalized histograms (bar) and their multimodal Gaussian fits (curve) of GPIbα (or GC) unfolding lengths pulled by engaged A1 (D,E) or AN51 (F,G) in indicated probe–target pairs. Peak values and percentages of unfolding lengths are indicated along with the identified unfolding domains. (H,I) Validation of MSD (H) and LRRD (I) unfolding. The WLC model was fit (curves) to the unfolding force vs. length data (black circles, mean ± s.e.m. of 15–25 measurements per point) from the WM23 vs. platelet experiments where only MSD unfolding was possible (H) or A1 vs. GC experiments where only LRRD unfolding was possible (I), yielding a contour length of 25.99 ± 0.85 nm or 70.29 ± 3.56 nm, respectively. Overlying on the two panels are corresponding unfolding force vs. length data (red triangles, mean ± s.e.m. of 20–30 measurements per point) from A1 vs. platelet ramped experiments where unfolding of MSD, LRRD or both were all possible, but were segregated into putative MSD (H) and LRRD (I) unfolding groups based on our decision rules in Figure 3—source data 1A. (J,K) Mean ± s.e.m. (n ≥ 20) of unfolding frequency (J) and length (K) of LRRD (A1 vs. GC) and MSD (WM23 vs. platelet). Force waveforms indicated as ramped force (1000 pN/s) and clamped force (25 pN) were generated with force-ramp and force-clamp experiment modes respectively. N.D. = Not detected.
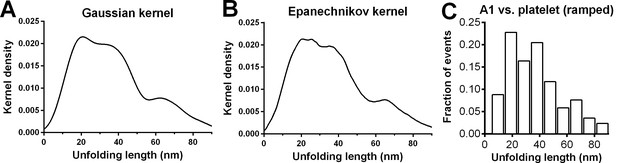
Statistical analysis on ramped unfolding length distribution.
(A,B) Nonparametric kernel density estimation using the software R with the given kernel ('gaussian' or 'epanechnikov') and bandwidth selected by Sheather and Jones (1991). Both estimates identify three populations in the distribution of the ramped unfolding length data from A1WT vs. platelet GPIbα experiments in Figure 2D. (C) Histogram analysis on the same data using the bin width 9.53 nm obtained using the Freedman-Diaconis formula and Freedman and Diaconis’ heuristic rule.
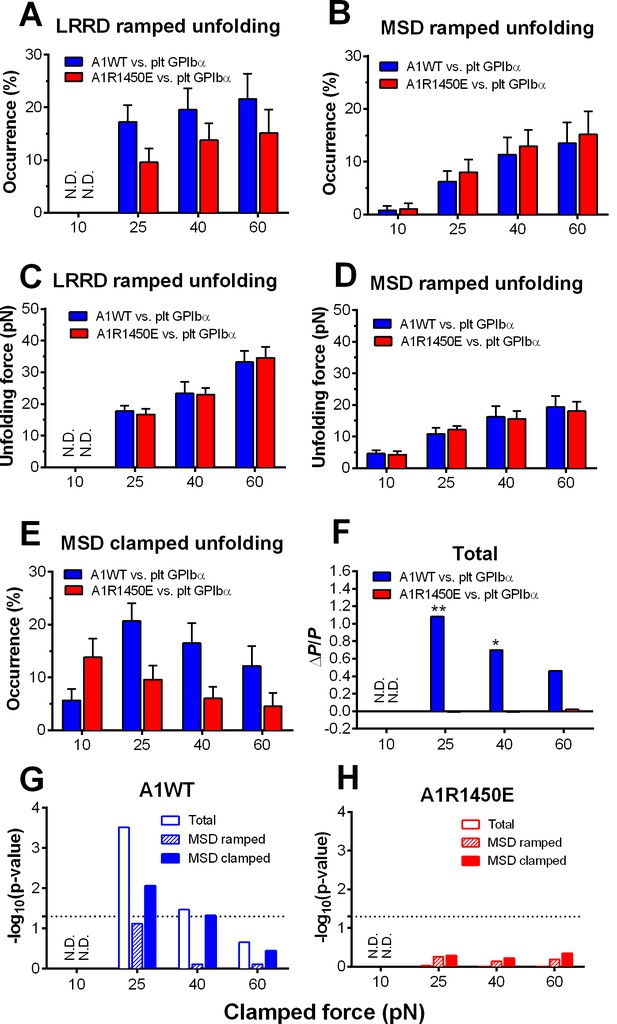
Force- and ligand-dependent cooperative unfolding of GPIbα LRRD and MSD.
(A–D) Frequency (A,B) and force (C,D) of LRRD (A,C) or MSD (B,D) unfolding events occurred in the ramping phase induced by pulling via A1WT (blue) or A1R1450E (red) with indicated preset clamped forces. (E) Occurrence frequencies of MSD clamped unfolding induced by holding at indicated clamped forces with A1WT or A1R1450E bonds. (F) The degree of cooperativity, quantified by ∆P/P = P(MSD+LRRD)/[P(MSD)×P(LRRD)] -1, is plotted vs. clamped force. P(LRRD), P(MSD) and P(LRRD+MSD) are the observed occurrence frequencies of unfolding events of LRRD alone, MSD alone and LRRD+MSD, respectively. (G,H) Significance of cooperativity assessed by (negative log10 of) p-value of the χ2 test of the null hypothesis H0: MSD unfolding and LRRD unfolding are independent. The χ2 test was not performed at 10 pN since under this force LRRD unfolding did not occur and hence no unfolding cooperativity. N.D. = not detected (A,C) or not done (F–H).
-
Figure 3—source data 1
Statistics and cooperativity evaluation of the GPIbα domains unfolding.
(A) Decision rules for and statistical summary of GPIbα domain unfolding in force-clamp experiment mode. Criteria for deciding whether or not (+ or −) and which (LRRD, MSD, or both) GPIbα domain(s) was (were) unfolded are based on BFP profile signatures and the unfolding lengths. YES = observed, NO = not observed. NA = not applicable. (B) Related to Figure 3F–H. Evaluation of LRRD and MSD unfolding cooperativity. All probabilities were calculated from occurrence data in (A). Observed joint probabilities were compared to their predicted counterparts based on the assumption that LRRD and MSD unfolded independently. For example, in 'WT A1 vs. Platelet' under 25 pN: The probability of LRRD unfolding is P(LRRD) = 3.4% + 6.9% + 2.76% = 13.06%. The probability of MSD unfolding is P(MSD) = 7.6% + 13.8% + 6.9% + 2.76% = 31.06%. The probability of MSD ramped unfolding is P(MSD, ramp) = 7.6% + 2.76% = 10.36%. The probability of MSD clamped unfolding is P(MSD, clamp) = 13.8% + 6.9% = 20.7%.
- https://doi.org/10.7554/eLife.15447.011
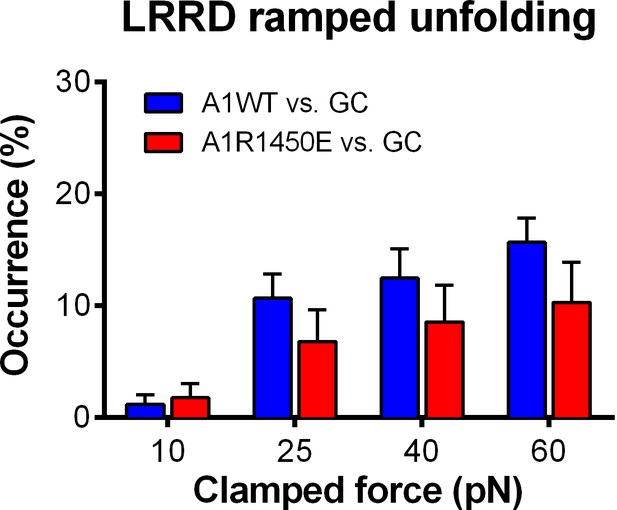
GC LRRD unfolding occurrence frequencies.
Frequencies of LRRD unfolding events occurred in the ramping phase induced by pulling GC via A1WT (blue) or A1R1450E (red) with 10, 25, 40 and 60 pN preset clamped forces.
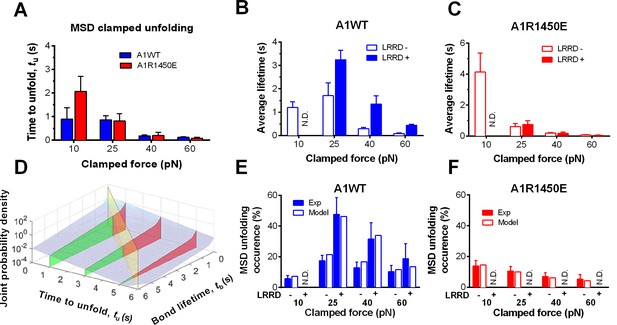
LRRD unfolding prolongs A1–GPIbα bond lifetime and facilitates MSD clamped unfolding.
(A–C) Mean ± s.e.m. of MSD time-to-unfold (tu, A) and GPIbα bond lifetimes (tb, B,C) with A1WT (blue) or A1R1450E (red) were measured in the clamping phase at different forces in the absence (−) or presence (+) of LRRD unfolding in the same BFP cycle. No LRRD unfolding occurred at 10 pN; hence no bond lifetime was measured under the LRRD+ at this force. (D) 3D plot of the surface of joint probability density (z-axis) of GPIbα to dissociate from A1WT at tb (x-axis) and MSD to unfold at tu (y-axis) (Materials and methods). Three planes, tu = 1, 3, and 5 s, under the probability density surface (gray) are shown in green or red, depending on whether they are on the left or right side of the tu = tb plane (yellow). (E,F) Measured (solid bars) and predicted (open bars) frequency of MSD unfolding events occurred in the clamping phase induced by the indicated force exerted via A1WT (E) or A1R1450E (F) in the presence (+) or absence (−) of LRRD unfolding in the same BFP cycle. N.D. = not detected. Error bar = s.e.m. estimated by the multinomial distribution of events.
-
Figure 4—source data 1
MSD unfolding rates (ku) and the fraction (w1) and off-rates (k1, k2) of GPIbα dissociating from A1WT or A1R1450E under different forces.
w1 represents the fraction of binding events that dissociate with the off-rate k1. The fraction of events that dissociate with the off-rate of k2 is simply calculated as w2 = 1-w1. NA = not applicable.
- https://doi.org/10.7554/eLife.15447.014
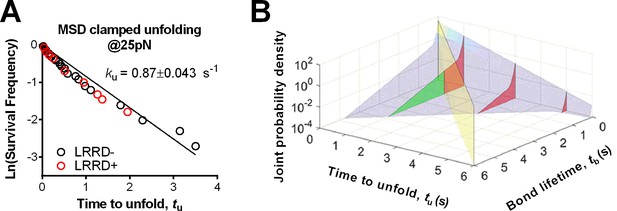
MSD time-to-unfold distribution for A1WT and 3D probability density surface plot for A1R1450E.
(A) Time-to-unfold (tu) distributions of A1WT MSD clamped unfolding with (red) and without (black) a preceding LRRD unfolding at 25 pN clamped force. The unfolding rate ku was calculated from the slope of ln(survival frequency) vs. time-to-unfold overlaid plot and the error were estimated from the 95% confident interval. (B) 3D plot of the surface of joint probability density (z-axis) of GPIbα to dissociate from A1R1450E at tb (x-axis) and the MSD to unfold at tu (y-axis) constructed based on Figure 4A,C (distributed ensemble data of A1R1450E at 25 pN and without LRRD unfolding).
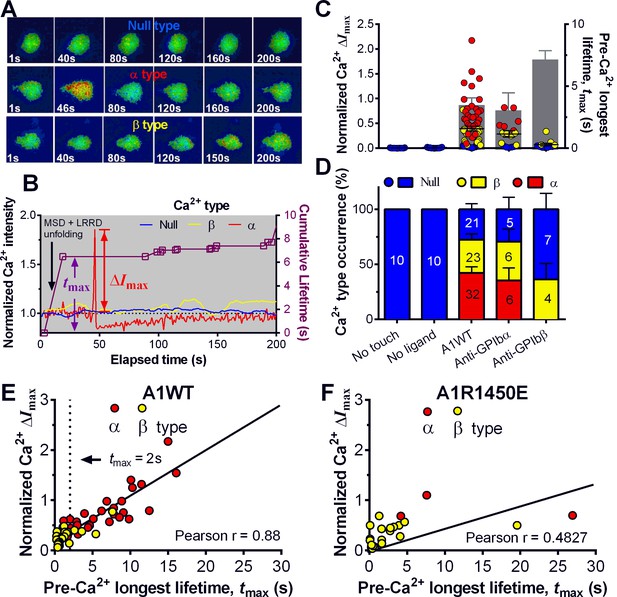
Concurrent analysis of single-platelet Ca2+ flux and GPIb-mediated single-bond binding at 25 pN clamped force.
(A) Representative epi-fluorescence pseudo-colored images of intraplatelet Ca2+ of null (top row), α- (middle row), and β- (bottom row) types at indicated times. (B) Representative time courses of normalized Ca2+ intensity of the null (blue), α (red) and β (yellow) types. The concurrent measurement of bond lifetime events (symbol) and the cumulative lifetime (curve) for the platelet exhibiting α-type Ca2+ is overlaid. The pre-Ca2+ longest lifetime (tmax) and the maximum intensity increase of the α-type Ca2+ (ΔImax) are indicated. The time when a concurrent LRRD and MSD unfolding event occurred is indicated by the arrow. (C,D) Individual ΔImax values and their mean ± s.e.m. (points, left ordinate) and mean ± s.e.m. of tmax (gray bars, right ordinate) (C) and fractions (D) of Ca2+ types triggered by different stimulations. Each point in (C) represents results from one platelet and the numbers of platelets in each column are indicated in the corresponding bar in (D), with matched colors to indicate Ca2+ types. (E,F) Scattergraphs of ΔImax vs. tmax for A1WT (E) and A1R1450E (F). The solid lines are linear fits to respective data with corresponding Pearson coefficients indicated. The null-type Ca2+ data was excluded in the analysis.
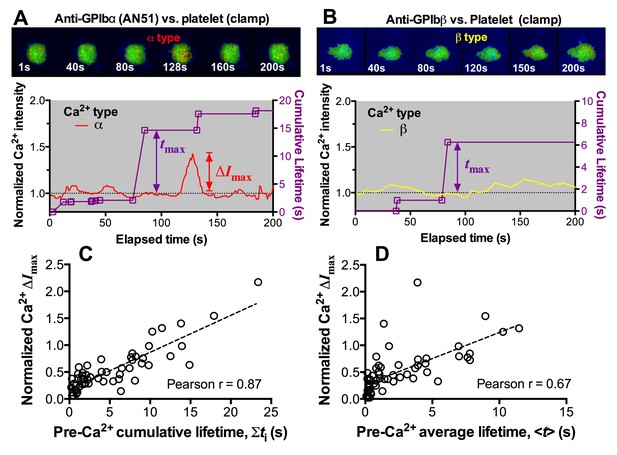
Concurrent analysis of single-platelet Ca2+ flux and single-bond dissociation from GPIb at 25 pN clamped force.
(A,B) Concurrent measurement of single-platelet Ca2+ flux and kinetics of single GPIb bonds with anti-GPIbα (A) and anti-GPIbβ (B). Top: representative epi-fluorescence pseudo-colored images of intracellular Ca2+ in platelets at indicated times. Bottom: Representative time courses of normalized intracellular Ca2+ intensity of the α- (A) and β- (B) types. The same keys are used as in Figure 5. (C,D) Normalized maximum Ca2+ intensity increase (ΔImax) vs. pre-Ca2+ cumulative bond lifetime ∑ti (C) or pre-Ca2+ pre-Ca2+ average lifetime <t> (D) of platelet GPIbα bonds with A1WT at 25 pN clamped force. Dashed lines are linear fits to data. The Pearson coefficients of the correlation (r) are indicated.
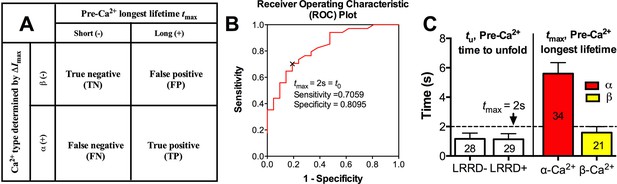
Specificity-sensitivity analysis of optimal threshold.
(A) Binary classifications of threshold performance outcomes on tmax–ΔImax data. (B) The receiver operating characteristic (ROC) plot: the true positive rate (i.e., sensitivity) against the false positive rate (i.e., 1 - specificity) at various tmax. The point of the optimal threshold t0 (tmax = 2 s) is identified by the black 'x', achieving the best sensitivity 0.7059 and specificity 0.8095. (C) Left: Mean ± s.e.m. of pre-Ca2+ MSD time-to-unfold (tu) measured from events with or without LRRD unfolding. Right: the tmax of platelets fluxing α- or β-type Ca2+. Dashed line indicates the tmax threshold that best distinguishes the two Ca2+ types.
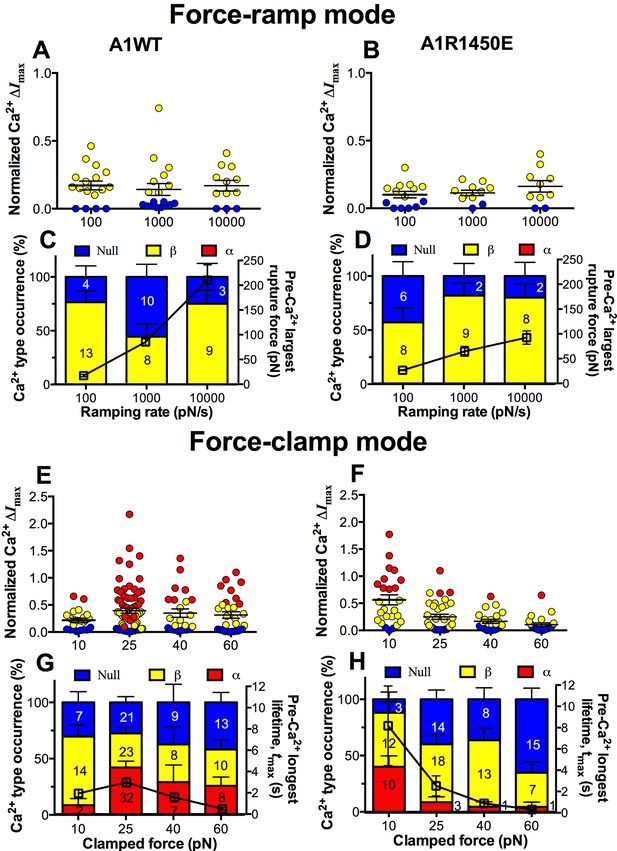
GPIbα can sense different force waveforms and discriminate different ligands.
(A–D) Force-ramp fBFP experiment mode. Individual ΔImax values and their mean ± s.e.m. (A,B, points), Ca2+ types (C,D, stacked bars, left ordinate), and mean ± s.e.m. of pre-Ca2+largest rupture force (C,D, black square, right ordinate) are plotted vs. force ramping rate for A1WT (A,C) or A1R1450E (B,D). (E–H) Force-clamp fBFP experiment mode. Individual ΔImax values and their mean ± s.e.m. (E,F, points), Ca2+ types (G,H, stacked bars, left ordinate), and mean ± s.e.m. of pre-Ca2+ longest lifetime (G,H, black square, right ordinate) are plotted vs. clamped force for A1WT (E,G) or A1R1450E (F,H). Each point in (A,B,E,F) represents results from one platelet and the numbers of platelets in each column are indicated in the corresponding bar in (C,D,G,H), with matched colors to indicate Ca2+ types. Error bar in (C,D,G,H) represents s.e.m. estimated by the multinomial distribution of events.
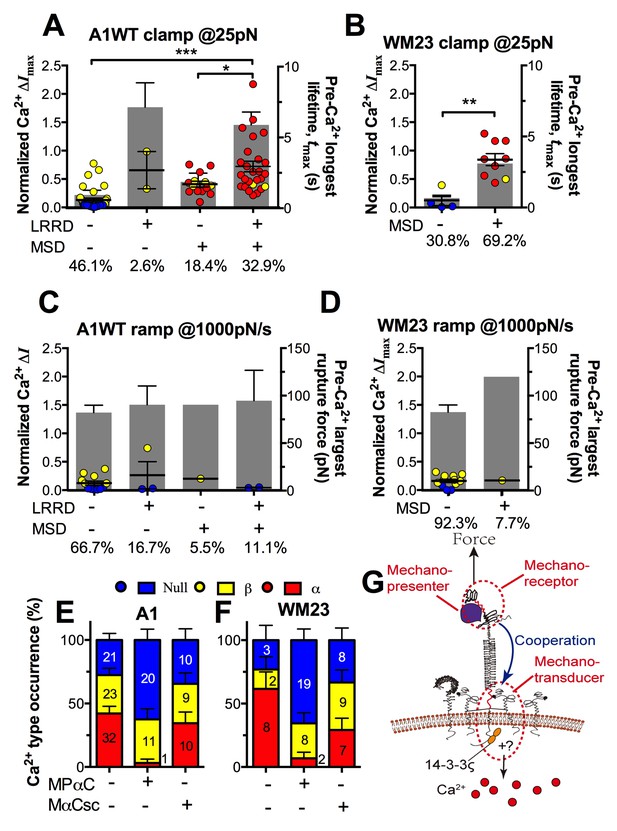
Correlation between GPIbα domain unfolding and Ca2+ triggering at 25 pN clamped force.
(A–D) Individual ΔImax values and their mean ± s.e.m. (points, left ordinate) in platelets triggered by A1 (A,C) or WM23 (B,D) binding, which were segregated into groups with (+) or without (−) unfolding of LRRD and/or MSD. Each point represents measurement from a platelet. The frequency of each unfolding combination to occur was indicated. (A,B) Data obtained from 25 pN force-clamp experiments. Corresponding tmax (gray bars, right ordinate) were overlaid with ΔImax. (C,D) Data obtained from 1000 pN/s force-ramp experiments. Corresponding pre-Ca2+ largest rupture force (gray bars, right ordinate) were overlaid with ΔImax. (E,F) Percentage of total events of three Ca2+ types in platelets in the same experiments as in (A) and (B) (left bars) as well as additional experiments performed in the presence of MPαC (middle bars) or MαCsc (right bars). Error bar = s.e.m. estimated from the multinomial distribution of events. (G) A postulated model of GPIbα-mediated mechanosensing. Force applied via VWF-A1 induces GPIbα LRRD and MSD unfolding. GPIbβ head domain binds to the unfolded MSD and causes the dissociation of its cytoplasmic tail from GPIbα-associated 14-3-3ζ, which transduces signals across the platelet membrane and further downstream, finally leading to α-type Ca2+. Each step of the mechanosensing process is indicated.
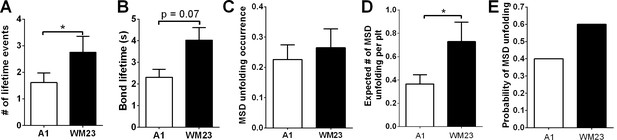
Comparison of GPIbα bond lifetime and MSD clamped unfolding by A1WT and WM23.
The number of lifetime events (A), their average lifetimes (B), MSD unfolding occurrence frequency (C), expected number of MSD unfolding events (D), and calculated probabilities of observing at least one MSD unfolding event (E) over a 200-s interrogation time during which a platelet was repeatedly contacted by a BFP bead coated with A1 or WM23. Data in panels A–C are presented as mean ± s.e.m. from n ≥ 60 events. The error bars in D are determined using Gaussian error propagation law. * indicates p < 0.05 by Student’s t-test.
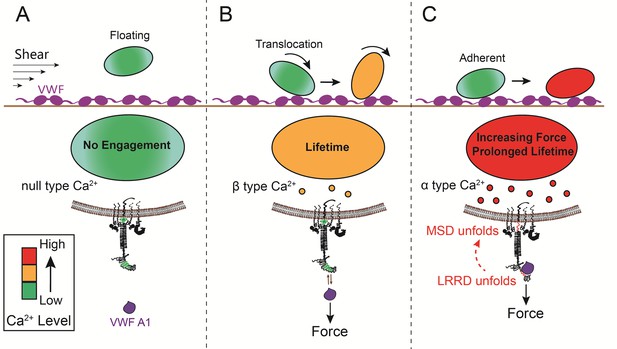
Model of GPIb-mediated platelet mechanosensing.
For a circulating platelet distal to an injured site without physical contact, its GPIb signaling will not be triggered (A). Upon interacting with VWF, platelets tether and translocate on the sub-endothelial surface via sequential intermittent VWF-A1–GPIbα bonds that provide adhesive forces. Having relatively short lifetimes, these forces unlikely induce MSD unfolding on GPIbα, hence only trigger β-type Ca2+ (B). LRRD unfolding under an increasing force prolongs bond lifetime and results in temporary platelet arrestment, providing a higher chance for MSD unfolding, which triggers α-type Ca2+ (C).
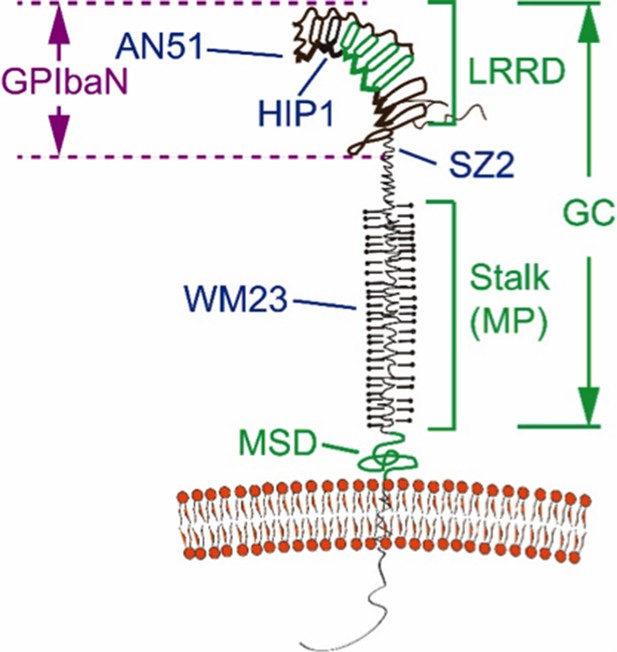
Domain organization and antibody epitopes on GPIbα.
https://doi.org/10.7554/eLife.15447.024Videos
BFP experiment mimics platelet translocation on sub-endothelium.
This animation (produced by Adobe Flash; 12 fps) illustrates the resemblance between platelet translocation on the sub-endothelium (a collagen network covered with VWF on the surface, upper panel) and the repetitive BFP experiment cycle (middle panel) with synchronized molecular interaction between a GPIbα and a VWF-A1 domain (lower panel). Two zoom-in platelet signaling scenarios are inserted following a short- and a long-lived VWF–GPIbα bond respectively. It starts with platelet translocation along the shear force direction, mimicked by the first two no-adhesion BFP cycles. An A1–GPIbα binding event with no lifetime in the BFP cycle results in a transient deceleration in the platelet translocation. After two more no-bond cycles, another bond event survives for a short lifetime without GPIbα unfolding. This triggers a signal (represented by purple stars) displaying a β-type Ca2+. Later, another bond event survives for a long lifetime, during which both LRRD and MSD unfold. This triggers a signal (represented by blue stars) displaying an α-type Ca2+.
Force-clamp experiment mode with a bond lifetime event.
The video consists of two parts in series. Part I is an animation (produced by Adobe Flash; 12 fps), and part II is a video recording of a representative fluorescence BFP experiment (recorded by a customized LabView program; 25 fps). Both parts show BFP force-clamp measurement cycles. In part I, the synchronized BFP illustration (upper panel), A1–GPIbα interaction (middle panel) and 'Force vs. Time' signal (lower panel) of the same force-clamp cycle with a lifetime event are displayed in parallel. Phases of the BFP cycle are indicated in the lower panel. Part II shows two BFP cycles, which sequentially render a no bond event and a bond lifetime event. The pseudo-color epifluorescence images (acquired at 1 fps) are interpolated and superimposed onto the brightfield images to reflect the real-time intraplatelet Ca2+ level (in a progressive sequence: blue, green, yellow, orange and red). Following the long lifetime event, calcium first rapidly elevates and then quickly decays, manifesting an α-type Ca2+.
Force-ramp experiment mode with a bond rupture event.
Similar to Video 2, this video consists of two parts in series. In part I, the synchronized BFP illustration (upper panel), A1–GPIbα molecular interaction (middle panel) and 'Force vs. Time' signal (lower panel) of the same force-ramp cycle with a ~65 pN rupture force event are displayed in parallel. Part II shows two BFP cycles, which sequentially render a no bond event and a bond rupture event. After the bond rupture event, low level calcium mobilization occurs right away, namely the β-type Ca2+.





