Phosphorylation acts positively and negatively to regulate MRTF-A subcellular localisation and activity
Figures
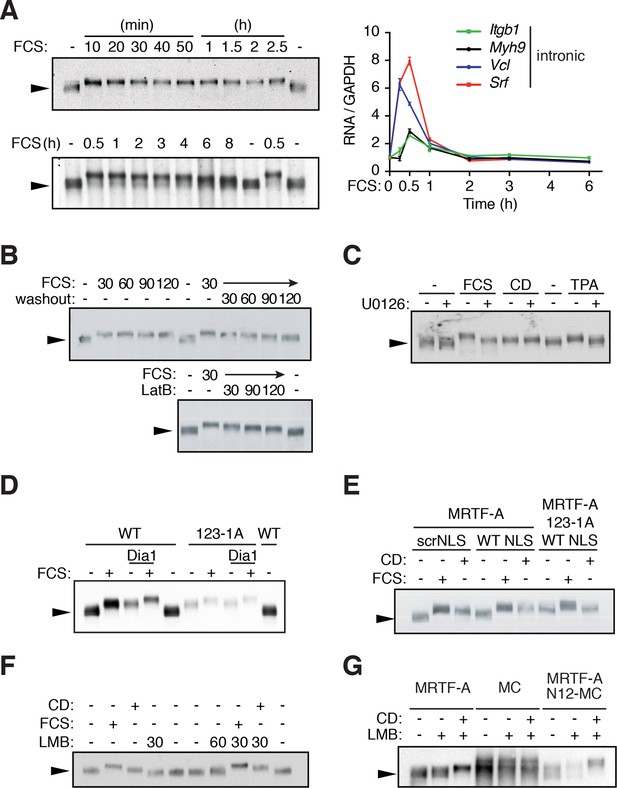
G-actin controls MRTF phosphorylation.
(A) MRTF-A phosphorylation persists after transcriptional shutoff. NIH3T3 cells were stimulated with 15% FCS for the indicated times. Left, Phosphorylation of MRTF-A (arrowed) was detected as reduced mobility in SDS-PAGE by immunoblot. Right, transcription of MRTF-A target genes, analysed by qPCR using intronic primers, normalised to GAPDH, and expressed relative to basal conditions. Data are from three independent experiments, plotted as mean ± SEM. (B) Cells were stimulated with 15% FCS as indicated; 30 min later the medium was replaced with 0.5% FCS or 1 μM Latrunculin B (LatB) was added, and endogenous MRTF-A phosphorylation monitored by SDS-PAGE. (C) Cells were stimulated with 15% FCS, 2 μM CD or 100 ng/ml TPA with and without 10 μM U0126. (D) MRTF-A phosphorylation is induced by mDia1 coexpression or disruption of G-actin binding. Cells expressing exogenous mouse MRTF-A or MRTF-A-123-1A, which cannot bind G-actin (Mouilleron et al., 2008; Vartiainen et al., 2007) and constitutively active mDia1 were treated with 15% FCS and MRTF-A phosphorylation monitored by SDS-PAGE. (E) Nuclear localisation of MRTF-A is not sufficient for its phosphorylation. Cells expressing MRTF-A linked to an intact or scrambled SV40 NLS were stimulated with 15% FCS or 2 μM CD for 30 min. (F) Cells were stimulated with 15% FCS, 2 μM CD or 50 nM Leptomycin B (LMB); MRTF-A phosphorylation was monitored by SDS-PAGE and immunoblotting. (G) Cells expressing MRTF-A, Myocardin (MC) or MRTF-A N12-MC, in which MC N-terminal sequences including RPEL1 and RPEL2 are replaced by those of MRTF-A (Guettler et al., 2008), were treated with 50 nM LMB with or without 2 μM CD as indicated. For effect on reporter gene activity, see Figure 1—figure supplement 1C.
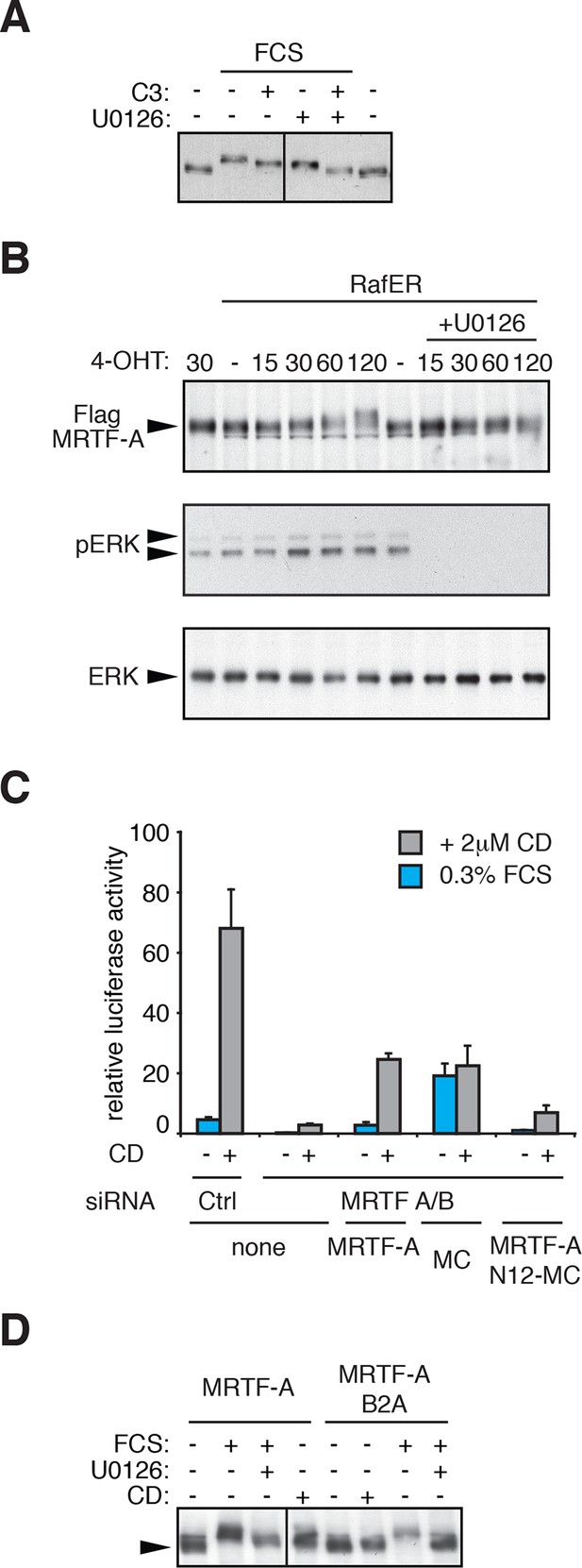
Regulation of MRTF-A phosphorylation.
(A) Cells were transfected with C3-transferase as indicated, maintained in media containing 0.3% FCS for 20 hr and then treated with 15% FCS and 10 μM U0126 as indicated. (B) Cells expressing MRTF-A and RafER were treated with 1 μM 4-Hydroxytamoxifen (4OHT) and 10 μM U0126 for the indicated times (min). MRTF-A (top), activated ERK (middle), and total ERK (bottom) were detected by immunoblotting. (C) Cells depleted of endogenous MRTFs were transfected with the MRTF-A-responsive 3DA-luc SRF reporter and plasmids encoding MRTF-A, Myocardin (MC) or MRTF-A N12-MC, in which MC N-terminal sequences including RPEL1 and RPEL2 are replaced by those of MRTF-A (Guettler et al., 2008) and treated with 2 μM CD as indicated. See also Figure 1G. (D) Cells expressing MRTF-A or MRTF-A B2A (KK152/154AA, Vartiainen et al., 2007) were treated with 15% FCS, 2 μM CD and 10 μM U0126 as indicated and MRTF-A phosphorylation monitored by SDS-PAGE and immunoblotting.
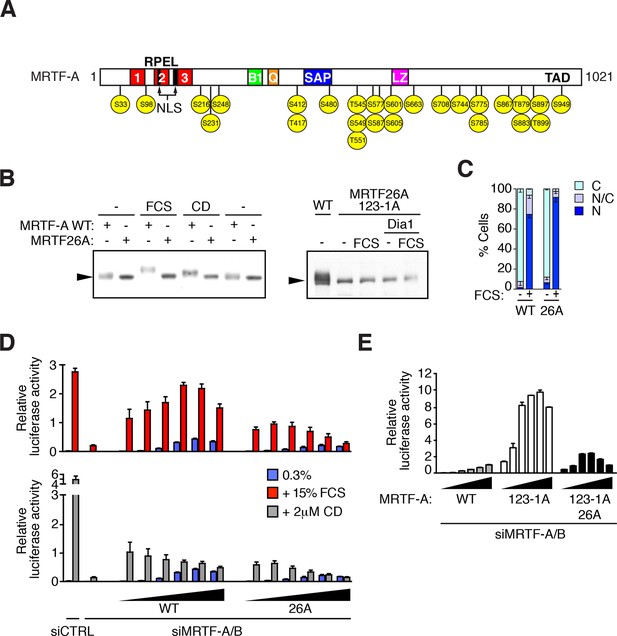
MRTF is phosphorylated at multiple sites.
(A) Schematic representation of MRTF-A, with S/T-P phosphorylation sites substituted with alanine in MRTF26A indicated in yellow. For details of phosphorylations, see Table 1 and Figure 2—figure supplement 1. (B) Phosphorylation is largely abolished in the MRTF26A mutant. Cells expressing wildtype MRTF-A or MRTF26A (left panel) or MRTF26A-123-1A (right panel), with or without activated mDia1, were stimulated with 15% FCS or 2 μM CD as indicated, and phosphorylation monitored by SDS-PAGE and immunoblotting. See Figure 2—figure supplement 2. (C) Cells expressing wild-type MRTF-A or MRTF-A 26A were stimulated with 15% FCS for 30 min. MRTF-A localisation was assessed by immunofluorescence (N, predominantly nuclear; N/C, pancellular; C, predominantly cytoplasmic), in two independent experiments. (D) Cells depleted of endogenous MRTFs were transfected with the MRTF-A responsive 3DA-luc SRF reporter and increasing amounts of wild-type MRTF-A or MRTF-A 26A (3, 6, 12, 25, 50, 100 ng). After treatment with 15% FCS or 2 μM CD, cell extracts were assessed for luciferase activity. Two independent experiments, data presented as mean ± half-range. (E) Cells were transfected with 3DA-luc and MRTF-A derivatives as in (D), and luciferase activity measured 24 hr later. Three independent experiments, data presented as mean ± SEM.
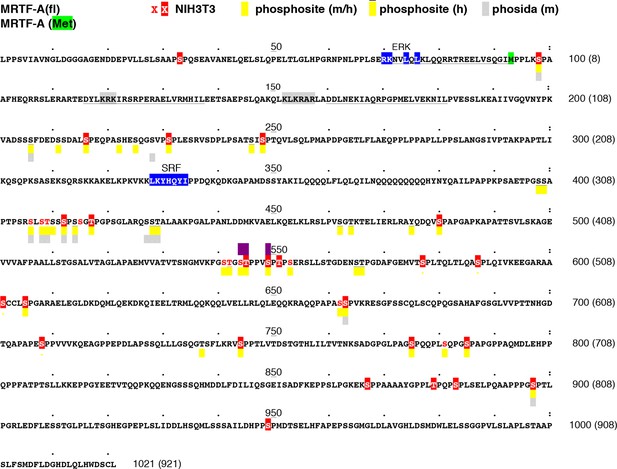
Location of MRTF-A phosphorylation sites.
The sequence of mouse MRTF-A(fl) is shown, with the initiation codon for MRTF-A(met) highlighted in green. RPEL motifs are underlined, the ERK- and SRF-binding sequences highlighted in blue, and the bipartite NLS highlighted in grey. Residues identified as phosphorylated in our analysis are in red; those substituted by alanine in MRTF-A26A are highlighted. Residues identified as phosphorylated in high-throughput analyses of mouse and human MRTF-A in the Phosphositeplus database are highlighted in yellow (human-only overlined) (http://www.phosphosite.org/proteinAction.action?id=15910&showAllSites=true/), and those in the Phosida database in grey (see http://141.61.102.18/phosida/index.aspx). Residues S544, T545 and S549, identified as putative ERK phosphorylation sites (Muehlich et al., 2008) and mutated to alanine in our mutant STS/A, are highlighted in purple. Sites S211, S216, S222, S225, S231, S406, S408, T409, S412, S414, S541, T542, T545, S549, S553, S605 and S662 have also been detected in MEFs by others (Hsu et al., 2011; Wu et al., 2012; Yu et al., 2011). Sites S541, T545, S549 and S553 correspond to a putative primed GSK3 phosphorylation site conserved in myocardin, which apparently negatively regulates its activity by promoting degration (Badorff et al., 2005; Xie et al., 2009). Sites S98, T545 and S549 have also been detected in EGF-stimulated HeLa cells (Olsen et al., 2006).
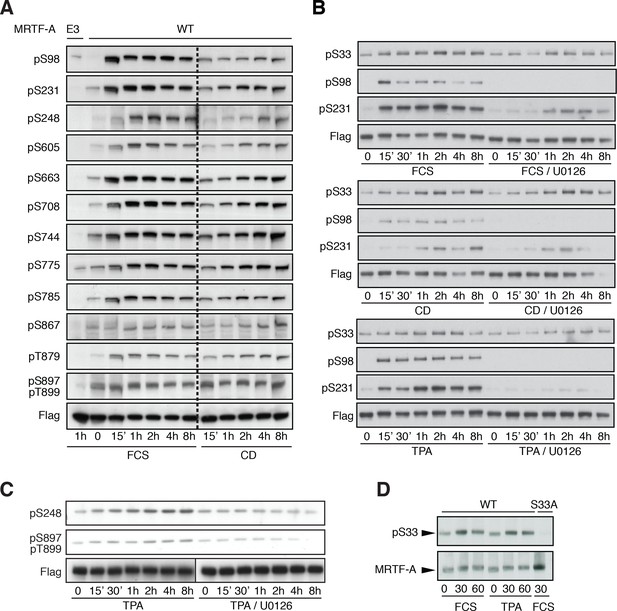
Detection of MRTF-A phosphorylation with phospho-specific antibodies.
(A) Cells were transfected with Flag-MRTF-A wild-type or mutant E3, in which sites including S98, S231, S248, S605, S663, S708, S744, S775, S785, S867, T879, S897 and T899 were substituted by alanine were stimulated with 15% serum or 2 μM cytochalasin D for the indicated times. Phosphorylation was detected by immunoblotting with the indicated phosphospecific antibodies. (B) Cells were transfected as in (A) and stimulated with 15% FCS, 2 μM cytochalasin D or 100 ng/ml TPA, in the presence of 10 μM U0126 as indicated before analysis with the indicated antibodies. (C) Cells were transfected as in (A) and stimulated with 100 ng/ml TPA, in the presence of 10 μM U0126 as indicated, before analysis with the indicated antibodies. (D) Cells transfected as in A, and analysed with anti-pS33 antiserum in the presence of unphosphorylated S33 peptide (SAAPSPQSEC).
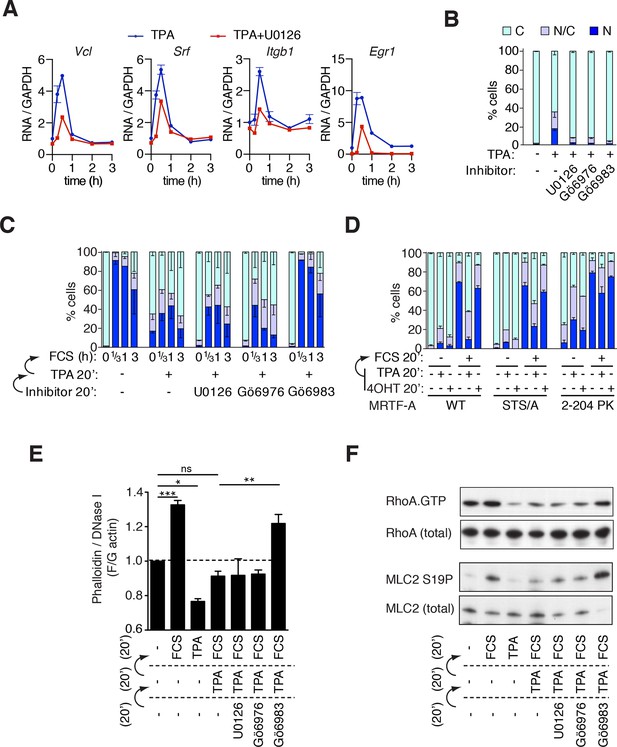
Phorbol esters act positively and negatively to control MRTF-A nuclear accumulation.
(A) TPA-induction of MRTF target genes. Cells were stimulated with 100 ng/ml TPA, and MRTF-A target gene pre-mRNA at 0.25, 0.5, 1, 2 and 3 hr analysed by qPCR, normalised to GAPDH and expressed relative to uninduced conditions. Data are mean ± half-range, n = 2. (B–D). Cells expressing Flag-MRTF-A or derivatives were pretreated with the indicated inhibitors for 20 min (U0126, 10 μM; Gö6976, 2 μM; Gö6983, 6 μM), then treated with 100 ng/ml TPA for 20', with serum stimulation for 20' as indicated. MRTF-A subcellular localisation was assessed by immunofluorescence. Data are mean ± half-range, n = 2. (B) TPA-induces MRTF-A nuclear accumulation (p<0.01, two-way ANOVA, Bonferroni post-test) but not in presence of inhibitors. (C) Cells as in (B) were stimulated with 15% FCS. Data are mean ± half-range, n = 2. Serum-induced MRTF-A nuclear accumulation was inhibited by pre-treatment with TPA (p<0.01), TPA+U0126 (p<0.01), and TPA+Gö6976 (p<0.05), but not TPA+Gö6983 (ns); two-way ANOVA with Bonferroni post-test. (Part B shows only the t = 0 points from this experiment for clarity). (D) Cells expressing Flag-MRTF-A, Flag-MRTF-A STS/A (S544A/T545A/S549A), or MRTF-A(2–204)-PK, together with Raf-ER as indicated, were pre-treated for 20' with 100 ng/ml TPA or 1 μM 4-hydroxytamoxifen (4OHT) then stimulated with 15% FCS for 20 min. Data are from two (WT and STS/A) or four independent experiments ((2–204)-PK). Only pre-treatment with TPA significantly blocked serum-induced nuclear accumulation (Predominantly nuclear MRTF-A cells: WT and STS/A, p<0.01; 2–204 PK, p<0.05; one-way ANOVA, Bonferroni multiple comparison test). (E) TPA inhibition of F-actin assembly in NIH3T3 cells is PKC-dependent but does not require ERK or conventional PKC. Cells were pretreated with the indicated inhibitors for 20 min, treated with 100 ng/ml TPA for 20 min, then stimulated with 15% FCS for 20 min, as in (C). After fixation F- and G-actin were quantified using fluorescently labelled phalloidin and DNase I, respectively, with F:G ratio in unstimulated cells set arbitrarily to unity. Plots are mean ± SEM, n = 3, with significance estimated by one-way ANOVA (*p<0.05; **p<0.01; ***p<0.001). (F) TPA inhibition of serum-induced RhoA activation and MLC2 phosphorylation is PKC-dependent but does not require ERK or conventional PKC. Lysates from cells treated as in panel (E) were used in a RhoA.GTP pull-down assay with GST-rhotekin as the affinity matrix (top); or analysed for MLC2 S19 phosphorylation by immunoblotting (bottom).
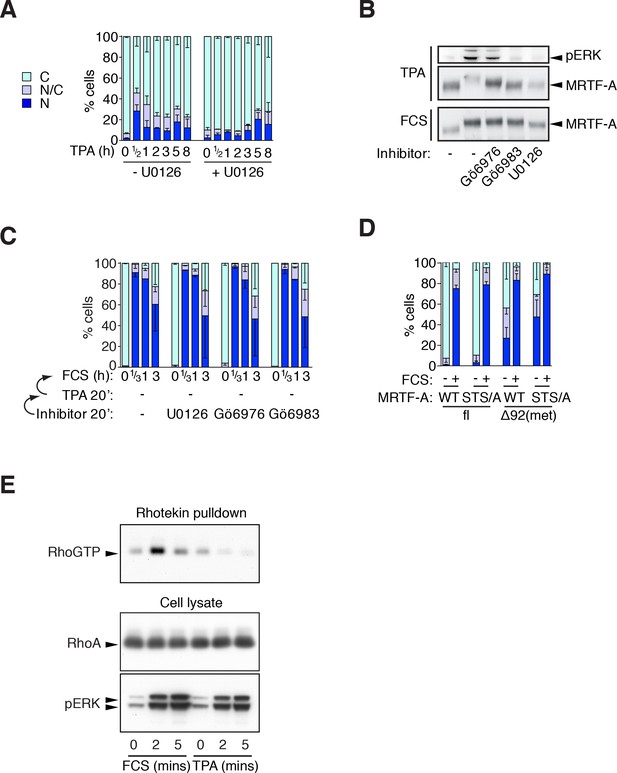
Phorbol esters act positively and negatively on MRTF-A nuclear accumulation.
(A) Cells expressing Flag-MRTF-A and maintained in 0.3% FCS were treated with 100 ng/ml TPA with or without 10 μM U0126 for various times. MRTF-A subcellular localisation assessed by immunofluorescence. N, predominantly nuclear; N/C, pancellular; C, predominantly cytoplasmic. Data are mean ± SEM, three independent experiments. (B) Cells expressing MRTF-A were pretreated for 20 min with U0126 (10 μM), Gö6976 (2 μM) or Gö6983 (6 μM), then treated with 100 ng/ml TPA or 15% FCS for 30 min before modification of MRTF-A and activation of ERK was analysed by immunoblotting. (C) ERK or PKC signalling is not required for serum-induced MRTF-A nuclear accumulation (controls for Figure 3C). Cells expressing Flag-MRTF-A were pretreated with inhibitors for 40' then stimulated with 15% FCS for the indicated times and MRTF-A subcellular localisation analysed. Data are mean ± half-range, two independent experiments. (D) Cells expressing Flag-MRTF-A ('fl') or Flag-Δ92MRTF-A ('met') with or without the STS/A (S544A/T545A/S549A) mutation, were stimulated with 15% FCS for 30 min and MRTF-A subcellular localisation was assessed as in (A). Data are mean ± SEM, three independent experiments. (E) Cells in 0.3% FCS were treated with 100 ng/ml TPA or 15% serum for the indicated times. RhoA.GTP pull-down assays were performed using GST-rhotekin as the affinity matrix and detection with RhoA antibody. Kinetics of ERK activation are compared in parallel by immunoblotting. See also Figure 3F.
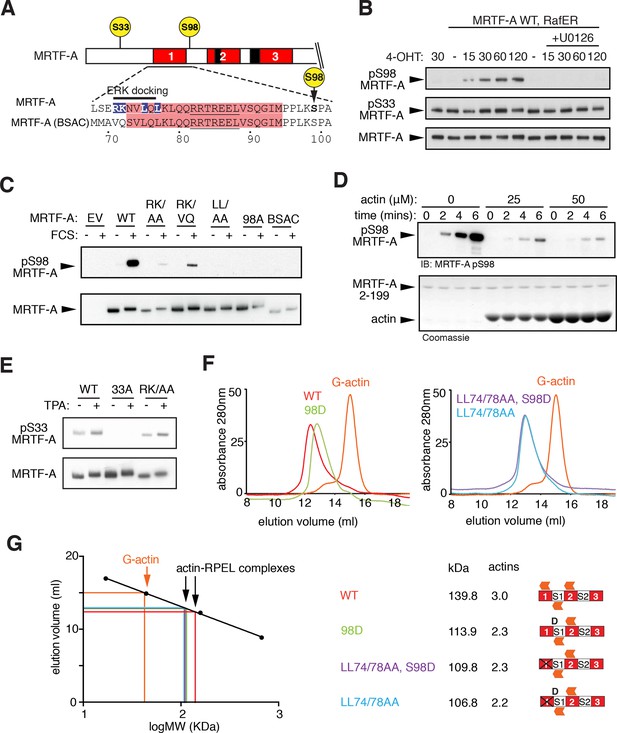
ERK-mediated phosphorylation of S98 inhibits actin binding.
(A) Sequences of MRTF-A(fl) and MRTF-A(BSAC), with RPEL1 highlighted in pink and the ERK-binding motif in blue. (B) Cells expressing Flag-MRTF-A and RafER were treated with 4OHT or vehicle and 10 μM U0126 as indicated. Lysates were analysed by immunoblot for MRTF-A pS98, MRTF-A pS33, and MRTF-A (Flag). (C) Cells expressing wild-type MRTF-A(fl) or derivatives with the indicated ERK-binding site mutations, or MRTF-A (BSAC), were analysed by immunoblotting with MRTF-A phospho-S98 antibody before and after serum stimulation. (D) Phosphorylation of recombinant MRTF-A(2-199) by ERK2 in the absence or presence of G-actin was monitored by MRTF-A phospho-S98 antibody (top) or coomassie staining (bottom). (E) S33 phosphorylation occurs independently of the ERK-binding motif. Cells expressing MRTF-A derivatives were analysed as in (C) using anti-MRTF-A phospho-S33. (F) Size exclusion chromatography of complexes formed between G-actin and recombinant MRTF-A(67–199) derivatives, or G-actin alone, colour coded. (G) Left, calibration curve; right, apparent stoichiometry of G-actin binding.
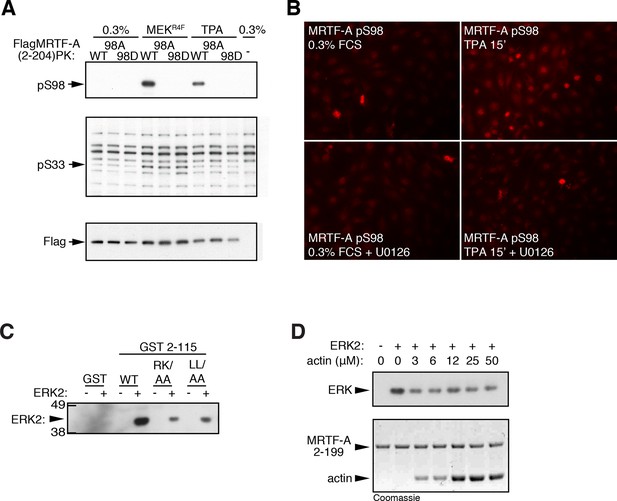
ERK phosphorylation of MRTF-A S98.
(A) Cells expressing Flag-MRTF-A(2–204)-PK and S98 mutants, with or without MEKR4F, were maintained in 0.3% FCS for 20 hr, then stimulated with 100 ng/ml TPA and phosphorylation of S98 and S33 analysed by immunoblotting. (B) Immunofluorescence assay of NIH3T3 cells before and after a 15' TPA stimulation with U0126 addition as indicated. (C) ERK interacts with the MRTF-A N-terminal sequences via the ERK docking motif. GST MRTF-A (2–115) or docking site mutant derivatives were used in pulldown assays with recombinant ERK2. (D) G-actin does not detectably compete for ERK binding in the pulldown assay. Assays with GST-MRTF-A (2–199) were as in (C) except that increasing amounts of G-actin were included.
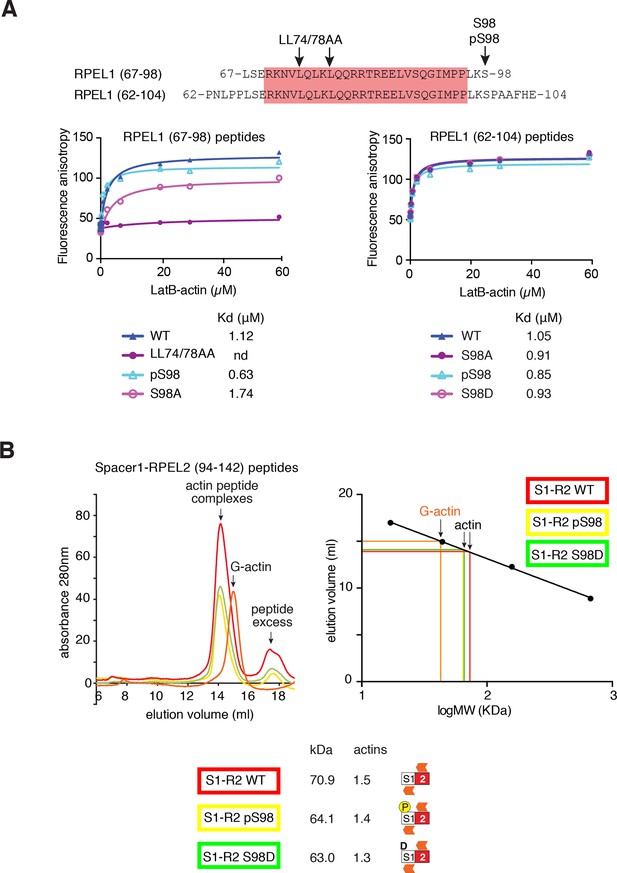
S98 phosphorylation impairs assembly of MRTF-A / G-actin complexes.
(A) Top, sequences of the RPEL1 containing peptides used in the fluorescence polarisation anisotropy assay, with mutation and phosphorylation sites indicated. Bottom, fluorescence anisotropy assay of G-actin binding to FAM-labeled RPEL1 peptides. Data represent mean of at least two independent experiments. (B) Size exclusion chromatography profiles of G-actin / Spacer1-RPEL2 peptide complexes or G-actin alone are shown, colour coded, with calibration at the right, and data are summarised below.
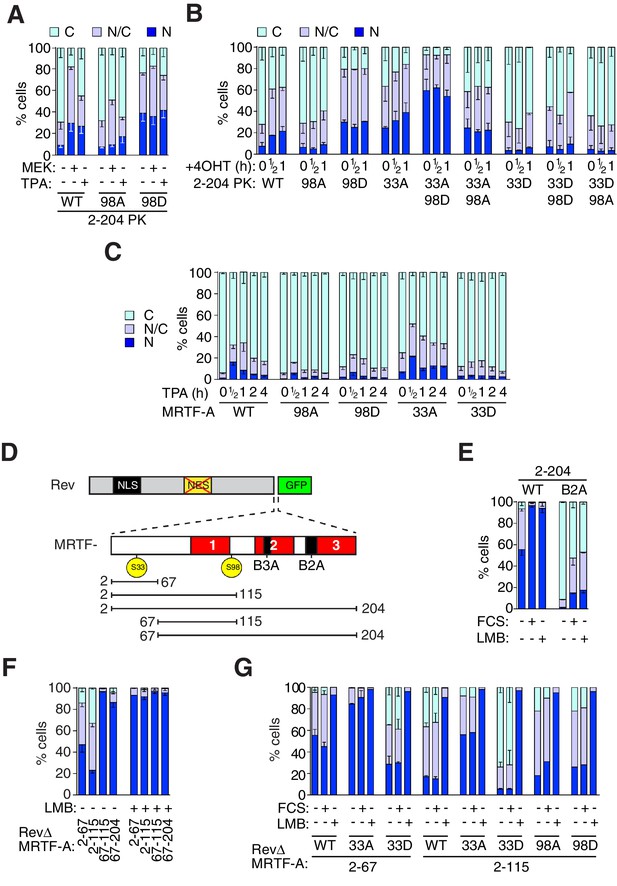
Phosphorylation of S98 and S33 has opposing effects on MRTF-A nuclear accumulation.
(A,B) Cells expressing MRTF-A derivatives, with or without activated MEK or RafER, were treated as indicated, and their subcellular localisation scored by immunofluorescence. (A) ERK-mediated S98 phosphorylation controls MRTF-A(2–204)PK localisation. Data are mean ± SEM, n = 3. Mutant S98D baseline higher than wildtype or S98A (p<0.01); MEK and TPA potentiate wildtype (p<0.001); 98A increased by MEK (p<0.01; possibly reflecting chronic ERK stimulation); others not significant. Two-way ANOVA with Bonferroni post-test. (B) Alanine substitutions or 'phosphomimetic' aspartate substitutions at S33 and S98 have opposing effects on MRTF-A(2–204)PK subcellular localisation. Data are mean ± half-range, n = 2. Mutant S98D baseline higher than wildtype or S98A (p<0.01); S98D and S33A cooperate to elevate baseline (p<0.001); 4OHT significantly induces wildtype and S33A (both p<0.05); two-way ANOVA with Bonferroni post-test. (C) N-terminal phosphosite mutations affect nucleocytoplasmic shuttling of intact MRTF-A. Data are mean ± half-range, n = 2. TPA treatment significantly increases nuclear localisation of wild-type MRTF-A (p<0.001), 33A (p<0.001) and 98D (p<0.05); one-way ANOVA, Bonferroni multiple comparison test. (D) Schematic representation of the MRTF-A / RevΔGFP derivatives. (E–G) analysis of MRTF-A NES activity by RevΔGFP assay. (E) MRTF-A(2–204) contains a Crm1-dependent NES. Data are mean ± half-range, n = 2. (F) MRTF-A residues 2–67, but not the RPEL domain, function as an NES. Data are mean ± SEM, n = 3. The 2–67 and 2–115 derivatives differ significantly from each other and from 67–115 and 67–204 in resting cells (all p<0.001 except 2–67 vs 67–204, p<0.01), but not following LMB treatment; two-way ANOVA with Bonferroni post-test. (G) Alanine or 'phosphomimetic' aspartate substitutions at S33, but not S98 affect MRTF-A N-terminal NES activity. Data are mean ± half-range, n = 2; absence of error bar shows a single datapoint (ie mean of two technical replicates).
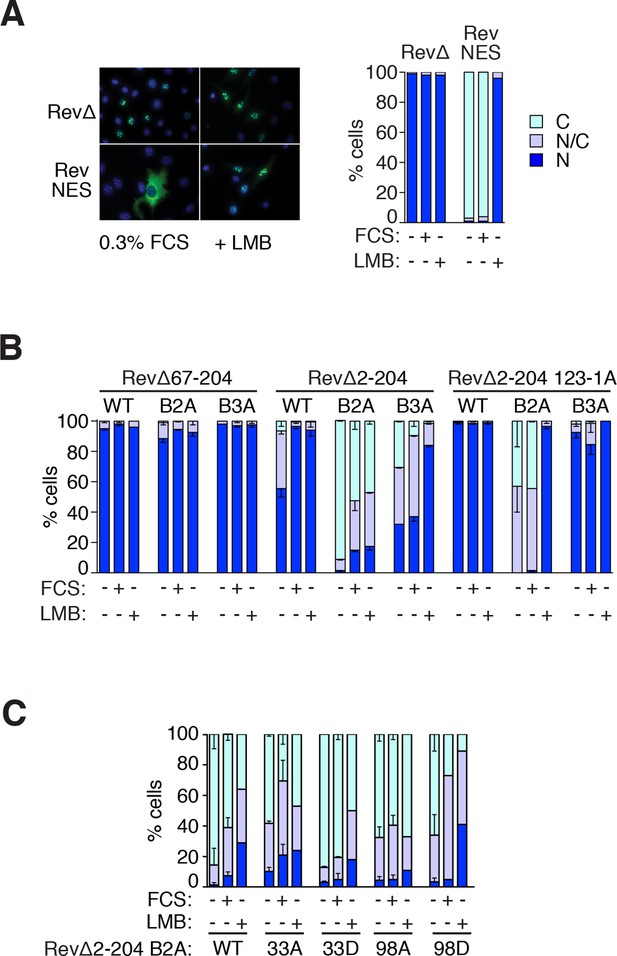
Use of RevΔGFP to detect NES activity in MRTF-A.
(A) The RevΔ nuclear export assay. Left, representative fluorescenbce images of RevΔGFP with or without re-insertion of the Rev NES and treatment with 50 nM LMB for 120 min. Right, serum does not induce relocalisation of RevΔGFP. N, predominantly nuclear; N/C, pancellular; C, predominantly cytoplasmic. A representative experiment is shown. (B, C) NES activity in the MRTF N-terminal sequences. N, predominantly nuclear; N/C, pancellular; C, predominantly cytoplasmic. Data are from two independent experiments. Error bars are half-range; absence of error bar shows a single point (i.e. mean of two technical replicates). (B) G-actin binding is required for the NES activity of the MRTF-A N-terminal sequences, but the RPEL domain does not exhibit NES activity. Each MRTF derivative was tested with weak (B3A) or strong (B2A) NLS mutations (Pawłowski et al., 2010) and scored as in (A). The MRTF-A RPEL domain (67–204) has no detectable NES activity even when the NLS is inactivated. Inclusion of N-terminal sequences reveals NES activity which is abolished by disruption of the G-actin-binding sites (123-1A mutant: alanine substituted at each RPEL motif). See also Figure 5. (C) Effect of alanine or 'phosphomimetic' aspartate substitutions at S33 and S98 on NES activity of RevΔMRTF-A(2–204) B2A, analysed as in (A).
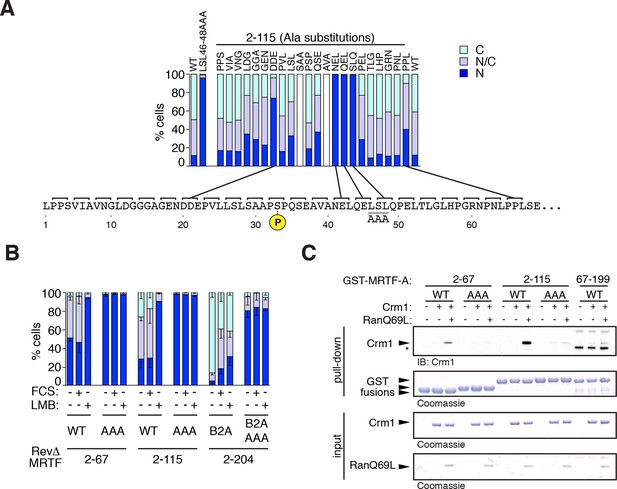
Mapping the Crm1-dependent N-terminal NES.
Subcellular localisation of RevΔGFP derivatives was scored by immunofluorescence. (A) Alanine-scan derivatives of RevΔ(2–115) GFP with the S33 phosphoacceptor and the LSL46-48AAA NES anchor-site mutations indicated. A representative experiment is shown. (B) Mutation of the NES core inactivates it. Data are mean ± SEM, n = 3. 2–67, 2–115, B2A(2–204) are significantly different from each other (all p<0.001); only B2A (2–204) exhibits significant change upon FCS stimulation (p<0.001); AAA mutants are significantly more nuclear than their parent (p<0.001); 2-way ANOVA with Bonferroni post-hoc test; 2–67/AAA comparison for N cells only). (C) The NES core is required for Ran-dependent Crm1 binding in vitro. Recombinant GST MRTF-A derivatives, with and without the NES anchor mutation, were used in pulldown assays with recombinant Crm1 and Ran 1–180 Q69L.
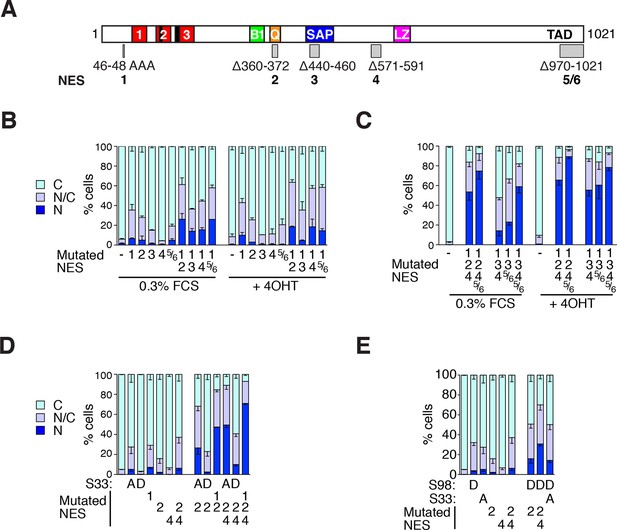
MRTF-A contains multiple cooperating NES elements.
Cells expressing MRTF-A derivatives, with or without RafER, were treated as indicated and their subcellular localisation was scored by immunofluorescence. (A) Summary of MRTF-A NES predictions and NES-inactivating deletions and point mutations (see also Figure 7—figure supplement 1A). (B–E) MRTF-A NES elements functionally cooperate with each other (B,C) and with N-terminal phosphorylations (D,E) to maintain cytoplasmic localisation of full-length MRTF-A. Data in panels D and E are from the same experiment. Data are mean ± half-range n = 2 (B,D,E) or mean ± SEM, n =3 (C). (B) NES1 cooperates with the C-terminal NES elements. Nuclear accumulation is enhanced by inactivation of NES1 (p<0.001), NES2 (p<0.001) or NES5/6 (p<0.05); inactivation of NES1 enhances the effect of inactivating NES2 and NES5/6 (p<0.001); one-way ANOVA with Bonferroni multiple comparison test. (C) Inactivation of NES5/6 significantly enhances effect of NES1/2/4 inactivation (p<0.001); one-way ANOVA with Bonferroni multiple comparison test. (D) Effect of alanine substitution at S33 is similar to NES1 inactivation. S33A enhances the effect of inactivation of NES2 (p<0.05) or NES2+4 (p<0.001), and is not significantly different from NES1 inactivation; NES1 inactivation enhances inactivation of NES2+NES4 (p<0.001); one-way ANOVA with Bonferroni multiple comparison test. (E) The S98D phosphomimetic mutation cooperates with inactivation of NES2 (p<0.01) and NES2+4 (p<0.001). One-way ANOVA, Bonferroni's multiple comparison test.
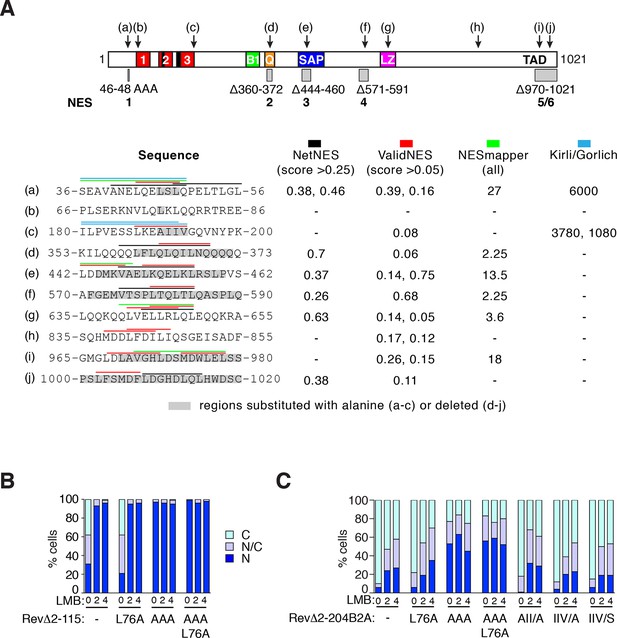
NES predictions and analysis.
(A) Prediction of NES elements in MRTF-A using online tools NetNES (la Cour et al., 2004), ValidNES (Fu et al., 2013), NESmapper (Kosugi et al., 2014) and an unpublished algorithm (K.Kirli and D. Gorlich, personal communication). Top, location of 10 potential MRTF-A NES elements (a–j) in MRTF-A, and the five NES-inactivating mutations used for functional analysis shown as grey boxes. Scores for the different elements are shown below, with coloured overlining indicating the sequences identified, and grey highlights indicating sequence mutated for functional analysis. Elements (b), (c), (g), and (h) were not studied further. (B) Analysis of putative NES (b) in the RevΔGFP assay, using RevΔ2–115. (C) Analysis of putative NES (b) and (c) in the RevΔGFP assay, using RevΔ2-204B2A. Data in (B) and (C) are from a representative experiment.
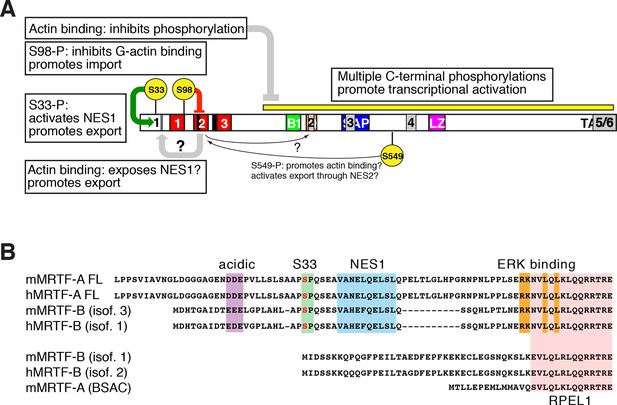
Roles of phosphorylation, NES elements, and actin binding in MRTF-A nucleocytoplasmic shuttling.
(A) Positive and negative roles of phosphorylation in MRTF-A, shown as in (Figure 7A) with the NES elements superposed in grey. G-actin binding inhibits MRTF-A phosphorylation, which is required for effective transcriptional activation. S98 phosphorylation inhibits formation of multimeric G-actin/MRTF complexes on the RPEL domain, promoting MRTF-A nuclear import; S33 phosphorylation potentiates the activity of NES1, which may be occluded in the absence of actin binding to the RPEL domain. The regulatory role of S549 phosphorylation in G-actin binding and NES2 activity (Muehlich et al., 2008), is indicated but is not detectable in our system. (B) Both MRTF-A and MRTF-B express variants containing or lacking the NES1 element and ERK docking site (Genbank: mouse MRTF-A FL, NP 694629.2 and AF532597.1; MRTF-A BSAC AAM 94258.1; MRTF-B isoform 3, NP 001116139.1; isoform 1, NP 705816.2, AAN 33042.1).
Tables
MRTF-A phosphorylation sites. NIH3T3 cells stably expressing Flag-MRTF-A were stimulated with serum for 30 min, immunoprecipitated with Flag antibody and fractionated by SDS-PAGE in triplicate. The MRTF-A bands were excised, digested with Trypsin, AspN or Chymotrypsin and the digests analysed with and without titanium dioxide enrichment on the LTQ Orbitrap Discovery. Fixed modifications were set as Carbamidomethyl (C) and variable modifications as Oxidation (M) and Phospho (STY). The estimated false discovery rate was set to 1% at the peptide, protein and site level. For all enzymatic digests, a maximum of two missed cleavages was allowed. Raw data were processed in MaxQuant (version 1.3.0.5) for peptide and protein identification, searching against the canonical sequence of Mus Musculus from the Uniprot KB, release 2012_08. The summary table was generated from the MaxQuant output file PhosphoSTY Sites.txt, an FDR-controlled site-based table compiled by MaxQuant from the relevant information about the identified peptides. Modified residues are shown in the left columns, with residue positions for MRTF-A(fl) and MRTF-A(met) indicated. In the sequence window the central residue is the putative modified site, shown in bold. For each site, the MS data are summarised as the best peptide Posterior Error Probability (PEP) score and the localisation probability, and/or the availability of a phospho-site specific antibody indicated. Blue shading indicates sites denoted reliable on the basis of their satisfying PEP <1.0E-06 and Localisation Probability >0.75 (>0.5 for repeated phosphoacceptors), confirmation by phosphosite-specific antibody, or manual peptide identification for residues 587 and 601 (peptides TQLTLQAS(ph)PL and AAS(ph)C(carb)C(carb)LS(ph)PGAR respectively, shown in blue in the sequence window). Residues mutated to Alanine in MRTF26A are shown in red. Peptides used for immunisation are underlined; those used for S98, S231, S663, S867 and ST897/9 were synthesised with C-terminal cysteines.
| Amino acid | MRTF-A(FL) | MRTF-A(Met) | Sequence | PEP score | Localizn prob. | Phospho-antibody |
|---|---|---|---|---|---|---|
| 33 | n/a | ENDDEPVLLSLSAAPSPQSEAVANELQELSL | 1.39E-10 | 0.993037 | pAb33 |
| 98 | 6 | TREELVSQGIMPPLKSPAAFHEQRRSLERAR | 1.16E-06 | 1 | pAb98 |
S | 206 | 114 | IIVGQVNYPKVADSSSFDEDSSDALSPEQPA | 0.0211238 | 0.310963 | |
S | 211 | 119 | VNYPKVADSSSFDEDSSDALSPEQPASHESQ | 0.000127434 | 0.799217 | |
S | 212 | 120 | NYPKVADSSSFDEDSSDALSPEQPASHESQG | 0.0059612 | 0.371831 | |
| 216 | 124 | VADSSSFDEDSSDALSPEQPASHESQGSVPS | 1.67E-30 | 1 | |
| 231 | 139 | SPEQPASHESQGSVPSPLESRVSDPLPSATS | 1.69E-23 | 1 | pAb231 |
| 248 | 156 | LESRVSDPLPSATSISPTQVLSQLPMAPDPG | 4.25E-16 | 0.999792 | pAb248 |
T | 402 | 310 | APPKPSAETPGSSAPTPSRSLSTSSSPSSGT | 0.000114318 | 0.993969 | |
S | 406 | 314 | PSAETPGSSAPTPSRSLSTSSSPSSGTPGPS | 9.19E-10 | 0.559961 | |
S | 408 | 316 | AETPGSSAPTPSRSLSTSSSPSSGTPGPSGL | 2.23E-11 | 0.676126 | |
T | 409 | 317 | ETPGSSAPTPSRSLSTSSSPSSGTPGPSGLA | 2.49E-09 | 0.497067 | |
| 412 | 320 | GSSAPTPSRSLSTSSSPSSGTPGPSGLARQS | 2.23E-11 | 0.99797 | |
S | 415 | 323 | APTPSRSLSTSSSPSSGTPGPSGLARQSSTA | 4.58E-06 | 0.524459 | |
| 417 | 325 | TPSRSLSTSSSPSSGTPGPSGLARQSSTALA | 2.34E-09 | 0.998496 | |
S | 428 | 336 | PSSGTPGPSGLARQSSTALAAKPGALPANLD | 0.000953078 | 0.745616 | |
T | 429 | 337 | SSGTPGPSGLARQSSTALAAKPGALPANLDD | 0.000953078 | 0.493371 | |
| 480 | 388 | KTELIERLRAYQDQVSPAPGAPKAPATTSVL | 2.99E-06 | 1 | |
S | 541 | 449 | MVVATVTSNGMVKFGSTGSTPPVSPTPSERS | 1.93E-11 | 0.96512 | |
T | 542 | 450 | VVATVTSNGMVKFGSTGSTPPVSPTPSERSL | 3.23E-07 | 0.899805 | |
S | 544 | 452 | ATVTSNGMVKFGSTGSTPPVSPTPSERSLLS | 3.78E-08 | 0.916768 | |
| 545 | 453 | TVTSNGMVKFGSTGSTPPVSPTPSERSLLST | 2.23E-12 | 0.994425 | |
| 549 | 457 | NGMVKFGSTGSTPPVSPTPSERSLLSTGDEN | 2.23E-12 | 0.999632 | |
| 551 | 459 | MVKFGSTGSTPPVSPTPSERSLLSTGDENST | 2.23E-12 | 0.94872 | |
S | 553 | 461 | KFGSTGSTPPVSPTPSERSLLSTGDENSTPG | 3.78E-08 | 0.998455 | |
S | 559 | 467 | STPPVSPTPSERSLLSTGDENSTPGDAFGEM | 0.00405521 | 0.999782 | |
T | 560 | 468 | TPPVSPTPSERSLLSTGDENSTPGDAFGEMV | 0.00405521 | 0.499997 | |
S | 565 | 473 | PTPSERSLLSTGDENSTPGDAFGEMVTSPLT | 0.000480998 | 0.837039 | |
| 577 | 485 | DENSTPGDAFGEMVTSPLTQLTLQASPLQIV | 5.33E-09 | 0.999998 | |
| 587 | 495 | GEMVTSPLQIVKEEGARAASC | 0.00523736 | 1 | |
| 601 | 509 | ASPLQIVKEEGARAELEGLD | 0.000780733 | 1 | |
| 605 | 513 | QIVKEEGARAASCCLSPGARAELEGLDKDQM | 2.27E-13 | 1 | pAb605 |
S | 662 | 570 | LQLEQQKRAQQPAPASSPVKRESGFSSCQLS | 1.02E-08 | 0.998348 | |
| 663 | 571 | QLEQQKRAQQPAPASSPVKRESGFSSCQLSC | 0.000148519 | 0.998905 | pAb663 |
| 708 | 616 | VPTTNHGDTQAPAPESPPVVVKQEAGPPEPD | 1.17E-24 | 1 | pAb708 |
| 744 | 652 | QLLLGSQGTSFLKRVSPPTLVTDSTGTHLIL | n/a | n/a | pAb744 |
| 775 | 683 | TVTNKSADGPGLPAGSPQQPLSQPGSPAPGP | 7.59E-16 | 1 | pAb775 |
S | 781 | 689 | ADGPGLPAGSPQQPLSQPGSPAPGPPAQMDL | 9.94E-13 | 0.998507 | |
| 785 | 693 | GLPAGSPQQPLSQPGSPAPGPPAQMDLEHPP | 3.03E-13 | 0.999999 | pAb785 |
| 867 | 775 | SADFKEPPSLPGKEKSPPAAAAYGPPLTPQP | 0.000807143 | 1 | pAb867 |
| 879 | 787 | KEKSPPAAAAYGPPLTPQPSPLSELPQAAPP | 0.0101532 | 1 | pAb879 |
| 883 | 791 | PPAAAAYGPPLTPQPSPLSELPQAAPPPGSP | 1.36E-08 | 1 | |
| 897 | 807 | PSPLSELPQAAPPPGSPTLPGRLEDFLESST | 1.92E-15 | 0.999573 | pAb897/9 |
| 899 | 809 | PLSELPQAAPPPGSPTLPGRLEDFLESSTGL | n/a | n/a | pAb897/9 |
S | 941 | 849 | PLSLIDDLHSQMLSSSAILDHPPSPMDTSEL | 0.000584126 | 0.569645 | |
| 949 | 857 | HSQMLSSSAILDHPPSPMDTSELHFAPEPSS | 8.45E-22 | 1 |





