Determination of ubiquitin fitness landscapes under different chemical stresses in a classroom setting
Figures
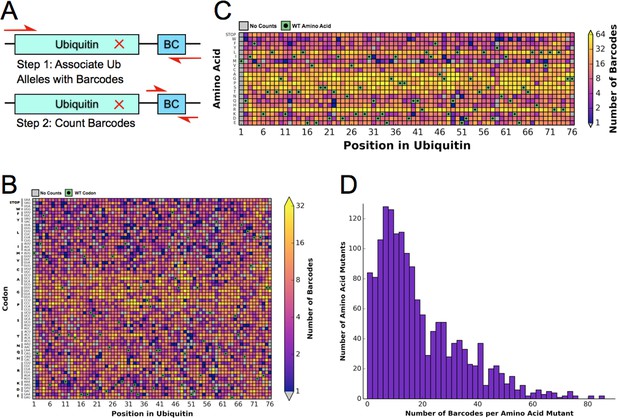
Barcoding enables a bulk competition experiment of ~1500 Ubiquitin variants.
(A) Prior to the competition experiment, ubiquitin alleles were specifically associated with unique barcodes through a paired end sequencing. To monitor the frequency of different alleles during the competition experiments, we directly sequenced the barcodes in a short single end read. (B) The library contains most codon substitutions and almost all are associated with multiple barcodes. A slight GC bias is seen in the cloning. WT codons are shown in green and missing alleles are shown in grey. (C) The amino acid coverage of the library is almost complete. WT residues are shown in green and missing alleles are shown in grey. (D) Examining the number of barcodes per amino acid substitution shows that 2.5% of the library is missing and the median number of barcodes per substitution is 15.
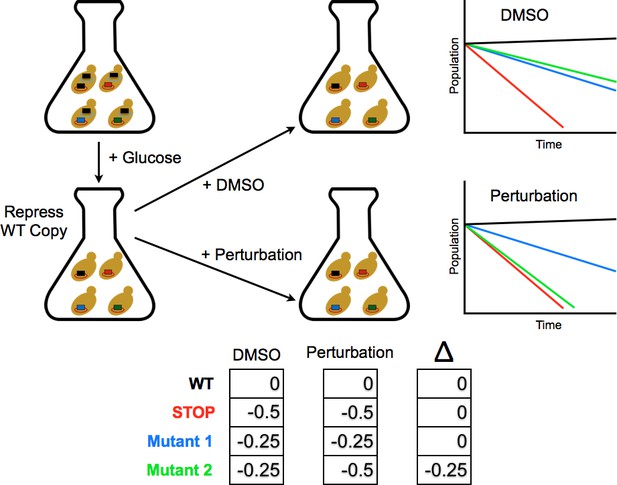
Competition experiment based on a galactose inducible Ub.
The fitness of all ubiquitin mutants was measured in a single culture by shutting off the galactose-driven wild type copy. This allows a constitutively expressed mutant to be the sole source of ubiquitin for the cell. The library was grown for 48 hr in galactose to remove dominant negative alleles and then expression of the wild type copy repressed by the addition of glucose. Upon repression of the wild type copy, chemical perturbations were added and the yeast were grown for multiple generations. Fitness scores were calculated for each mutant based on the relative frequencies of mutant and wild type alleles over multiple generations. The ratio of (mutant counts):(wild type counts) was computed for each time point and a line fit to these ratios vs. generation time. The fitness score is the slope of the linear fit.
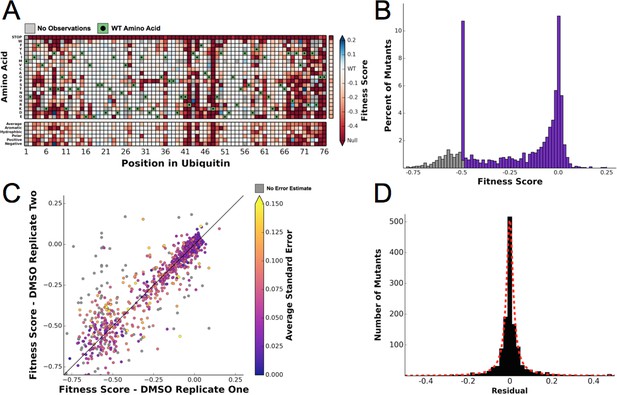
Ubiquitin fitness scores determined in DMSO are replicable and define the 'unperturbed' Ub fitness landscape.
(A) Heatmap showing the fitness of observed ubiquitin alleles. Scores presented are the average of three biological replicates. Wild type amino acids are shown in green and mutations without fitness values (due to lack of barcode or competition sequencing reads) are shown in grey. The average fitness score of each position and the averages of substitutions binned by amino acid characteristics are shown below. The single column on the far right shows the average of each amino acid substitution across all positions. (B) The distribution of fitness values is shown and colored based on fitness score. Grey bins reflect fitness scores that were reset to -0.5. (C) Biological replicates of the competition experiment in DMSO are well correlated (R2 = 0.79). Each point represents the fitness score of a mutant in two biological replicates. Points are colored based on the average standard deviation of the barcodes contributing to each fitness score. (D) The distribution of the residuals to the identity line between two DMSO replicates is symmetric and well modeled by a Lorentzian (X0 = 0, Γ = 0.0035, scaled by 1600).
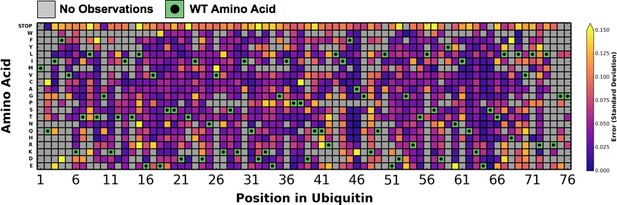
Error estimates for fitness scores determined in DMSO.
We calculated the standard deviation of the distributions of barcode fitness scores that contribute to each amino acid mutant fitness score. Large errors of the stop codon substitutions are due to variation of fitness scores below the -0.5 floor.
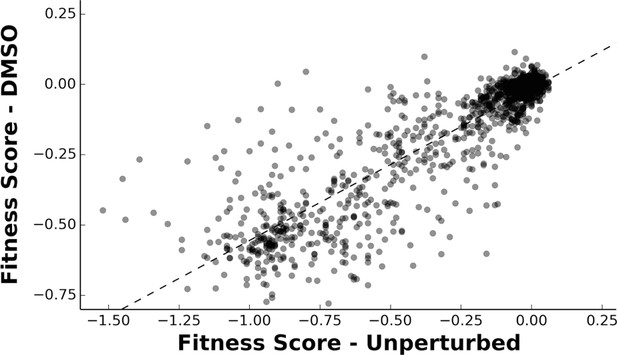
Fitnesses determined in DMSO are well correlated to the previously determined unperturbed fitnesses.
A linear regression (R2 = 0.785) and Pearson’s correlation coefficient (CC = 0.886) were calculated between the fitness scores determined in DMSO and the previously published unperturbed dataset (Roscoe et al., 2013).
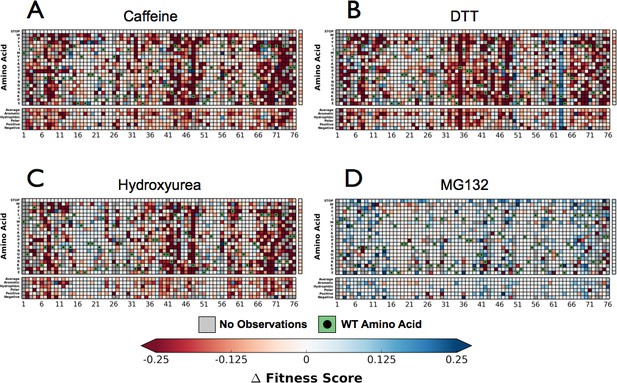
Perturbations sensitize ubiquitin to mutations.
The difference in fitness between DMSO and a perturbation for each Ub allele: (A) Caffeine, (B) DTT (C) Hydroxyurea (D) MG132. Wild type amino acids are shown in red and mutations without fitness values (due to lack of barcode or competition sequencing reads) are shown in grey.
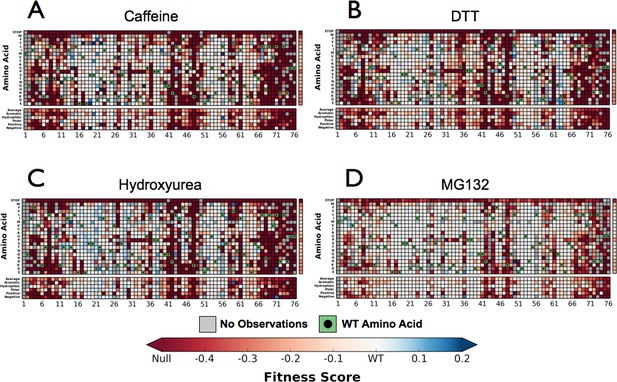
Perturbations sensitize ubiquitin to mutations.
Heatmaps showing the fitness of observed Ub alleles under: (A) Caffeine, (B) DTT (C) Hydroxyurea (D) MG132. Wild type amino acids are shown in green and mutations without fitness values (due to lack of barcode or competition sequencing reads) are shown in grey.
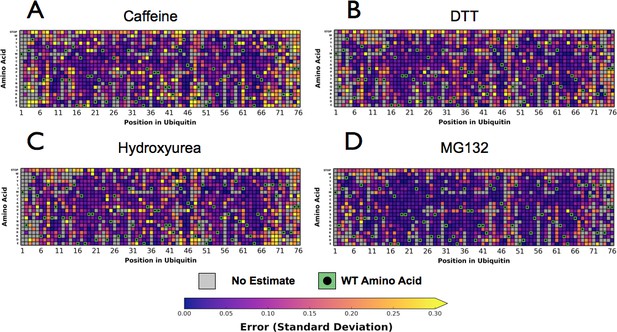
Perturbations sensitize ubiquitin to mutations.
Heatmaps showing the error estimates for each observed Ub alleles under; (A) Caffeine, (B) DTT (C) Hydroxyurea (D) MG132. Wild type amino acids are shown in green and mutations without fitness values (due to lack of barcode or competition sequencing reads) are shown in grey. Growth rates were calculated by monitoring OD600 over 8 hr and normalized to the unperturbed SUB328 growth rate.
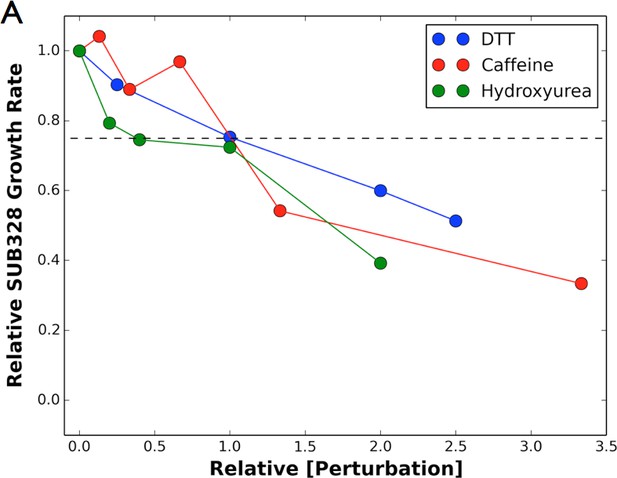
Growth curves.
We determined the concentration to inhibit SUB328 growth by 25% by monitoring optical density. Error bars represent standard deviation of multiple measurements.
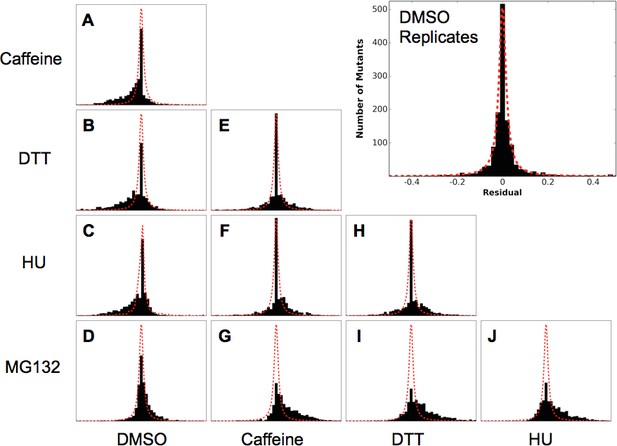
Residual distributions highlight a shared mutational response between Caffeine, DTT and HU.
The residuals between datasets shows are shown with the Lorentzian representing the biological replicates of DMSO in red. When compared to DMSO, three perturbations (Caffeine, DTT and HU) shift the distributions to the left, which highlights the increased sensitivity to mutation. In contrast, MG132 slightly shifts the distribution to the right, which highlights the alleviating interaction between MG132 and deleterious ubiquitin alleles. Comparisons between Caffeine, Hydroxyurea and DTT are symmetric but with longer tails than the control experiments. This result suggests a shared response comprised of many sensitized residues and a smaller number of perturbation-specific signals.
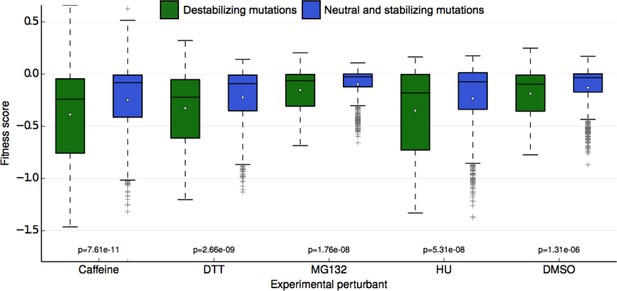
Fitness score data binned by Rosetta stability predictions.
Fitness scores for each of the 5 sets of experimental conditions are shown along the y-axis as boxplots. Scores are grouped first by their respective experimental condition, and then by the change in stability of the ubiquitin monomer of the mutation estimated by Rosetta. Mutations that Rosetta predicts to be neutral or stabilizing (REU (Rosetta Energy Units) < 1.0) are shown in blue boxes; mutations predicted to be destabilizing (REU >= 1.0) are shown in green boxes. The mean of each fitness score distribution is shown as a white dot. The p-value of the two-sided T test between the fitness mean of mutations predicted to be stabilizing and those predicted to be neutral/stabilizing is shown at the bottom of the plot. Experimental conditions are arranged from left to right along the x-axis in order of decreasing p-value.
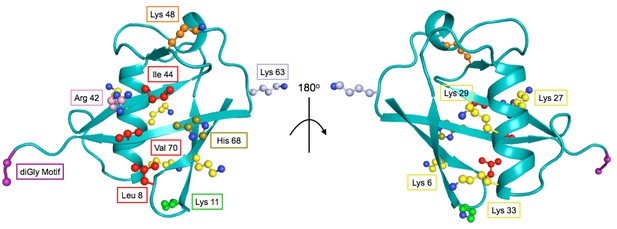
The structure of Ub highlighting important residues.
Cartoon model of Ub (PDB 1UBQ) with important residues colored as follows: Lys48 - orange, Lys63 - light blue, Lys11 - green, other Lys residues - yellow, hydrophobic patch (Leu8, Val40, Ile 44) - red, C-terminal diGly motif (Gly75 and 76) - purple, Arg42 - pink, His68 - olive.
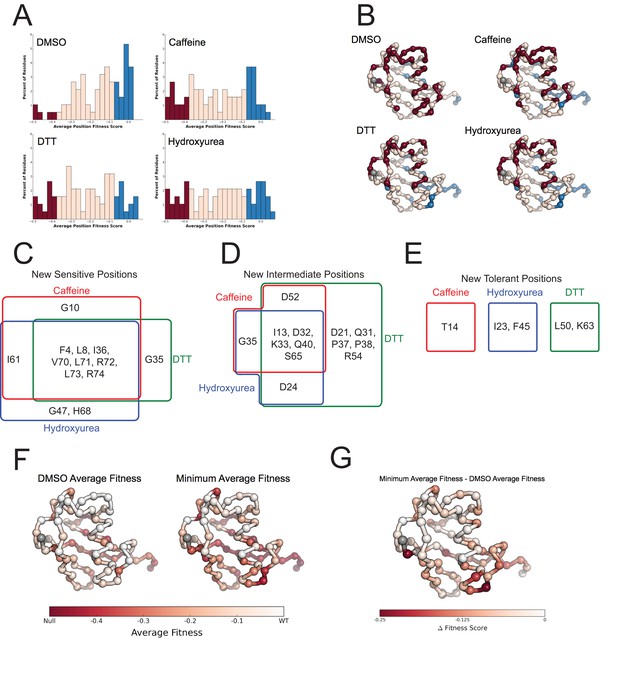
Average fitness values show sensitization by the perturbations at each position in ubiquitin.
(A) Based on the average fitness score, positions were binned into tolerant (>=-0.075 - Blue), intermediate (<-0.075 to > -0.35 - Pink) and sensitive (<= -0.35 - Red). (i) DMSO (ii) Caffeine (iii) DTT (iv) Hydroxyurea show a shift from tolerant to intermediate and sensitive positions. (B) Positions binned by average fitness score mapped onto the ubiquitin structure. C-alpha atoms are shown in spheres and the residues are colored as in A. Met1 is colored grey. (C) New sensitive positions induced by the perturbation describe a shared response to perturbation with 8 of 13 positions shared between Caffeine, DTT and HU. (D) New intermediate positions highlight the similarity between HU and Caffeine, with DTT sensitizing a unique set of residues. (E) New tolerant positions are unique to each perturbation. (F) Average position fitness scores mapped onto ubiquitin. (i) DMSO (ii) Minimum average fitness score in all perturbations. C-alpha atoms are shown in spheres and the residues are colored according to average fitness. Met1 is colored grey. (G) Minimum average fitness scores – DMSO average fitness scores mapped onto ubiquitin. C-alpha atoms are shown in spheres and the residues are colored according to the difference in fitness. Met1 is colored grey. With this small set of perturbations most positions are sensitized.
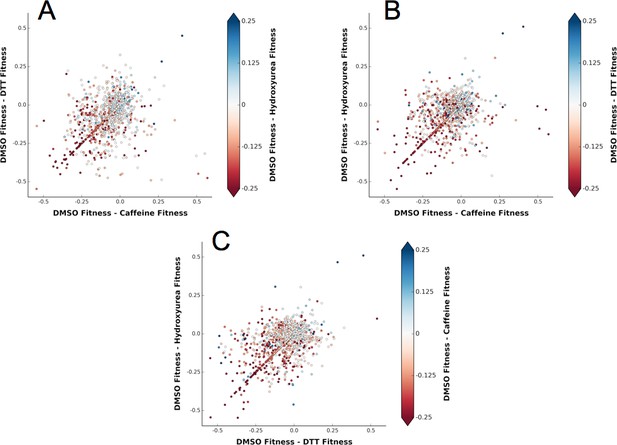
A shared response to different chemical perturbations.
(A) DMSO fitness - Caffeine fitness vs. DMSO fitness - DTT fitness. The markers are colored based on DMSO fitness - Hydroxyurea fitness. (B) DMSO fitness - Caffeine fitness vs. DMSO fitness - Hydroxyurea fitness. The markers are colored based on DMSO fitness - DTT fitness. (C) DMSO fitness - DTT fitness vs. DMSO fitness - Hydroxyurea fitness. The markers are colored based on DMSO fitness - Caffeine fitness.
-
Figure 9—source data 1
Shared response mutants representing mutations that are equally perturbed by all three sensitizing perturbations.
Mutants in the shared response were determined by fitting a line to the fitness scores. The distance from each point to that line was calculated. If the distance was less than 0.1 and the average Δ (DMSO - Perturbation) fitness was less than -0.2 the mutant was considered part of the shared response. E1 activity relative to WT Ub (Roscoe and Bolon, 2014) is listed and may explain the sensitization of some of the shared response mutants.
- https://doi.org/10.7554/eLife.15802.017
-
Figure 9—source data 2
Perturbation specific mutations represent alleles that are differentially affected by Caffeine, DTT and Hydroxyurea.
Perturbation specific mutations were determined by fitting a line to the delta (DMSO - perturbation) fitness scores. The distance from each point to that line was calculated. If the distance was greater than 0.35 the mutant was classified as perturbation specific. Mutants with high experimental errors were deemed outliers and removed from this list.
- https://doi.org/10.7554/eLife.15802.018
-
Figure 9—source data 3
Specific information regarding highlighted mutants.
- https://doi.org/10.7554/eLife.15802.019





