An inhibitory corticostriatal pathway
Figures
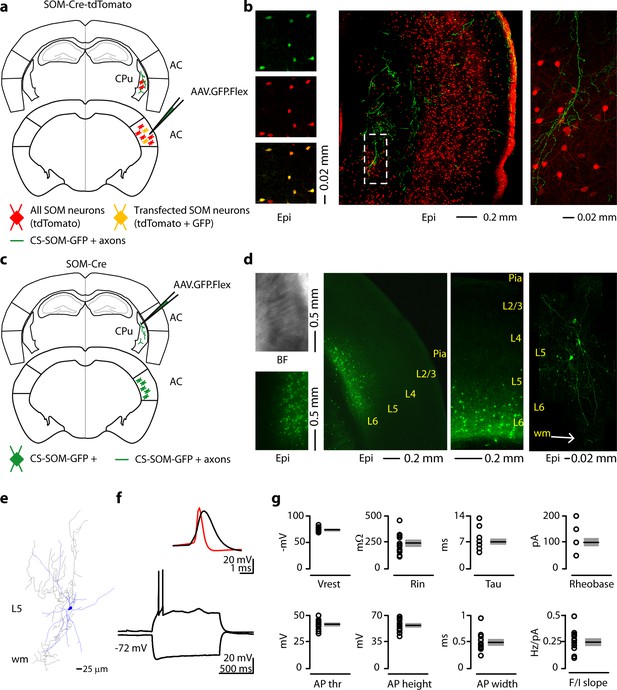
Morphological characteristics, axonal projections and electrical properties of long-range CS-SOM neurons in the mouse auditory cortex.
(a) Schematic depicting injection site using the SOM-Cre-tdTomato transgenic mouse line to identify CS-SOM neurons and their projections to the dorsal striatum. Bottom, auditory cortex: AAV.GFP.Flex injection site; yellow CS-SOM somata coexpressing GFP and tdTomato. Top, dorsal striatum: green CS-SOM GFP-positive axons; red SOM tdTomato-positive interneurons. (b) Epifluorescence images of SOM GFP-positive neurons. Top, left: GFP-positive SOM neurons in the auditory cortex identified by viral injection of AAV.GFP.Flex in the SOM-Cre-tdTomato transgenic mouse line. Middle, left: tdTomato-expressing SOM neurons in the SOM-Cre-tdTomato transgenic mouse line. Bottom, left: overlay of GFP and tdTomato images. Middle, the dashed box indicates the location GFP-positive axons from CS-SOM neurons in the dorsal striatum and the location of image in the right panel. Right, higher magnification of GFP fluorescence of CS-SOM axons in the dorsal striatum. (c) Schematic depicting injection site using the SOM-Cre transgenic mouse line to identify CS-SOM neurons by anatomical retrograde transfection. Top, striatum: AAV.GFP.Flex injection site. Bottom, auditory cortex: green CS-SOM GFP positive somata. (d) Bright-field (top left) and epifluorescence (bottom left) images of striatal SOM interneurons transfected with AAV.GFP.Flex. Middle (left and right), epifluorescence image of laminar distribution of CS-SOM neurons in the auditory cortex identified by anatomical retrograde transfection. Right, high-resolution image of a biocytin-labeled CS-SOM neuron. (e) Morphological reconstruction of one CS-SOM neuron (dendrites, blue; axons, gray). (f) Bottom, train of action potentials recorded in a GFP-positive CS-SOM neuron during step current injection (1.0 s, 100 pA pulse). Top, single action potential from GFP-positive CS-SOM neuron (black); compare to an action potential from a fast-spiking interneuron (red). (g) Summary plot of Vrest: resting membrane potential; Ri: input resistance; Tau: membrane time constant; Rheobase, the smallest current step evoking an action potential; AP thr: action potential threshold; AP height: action potential height; AP half-width: action potential half-width; and F/I slope from CS-SOM neurons (n = 13), including group averages (± s.e.m.).
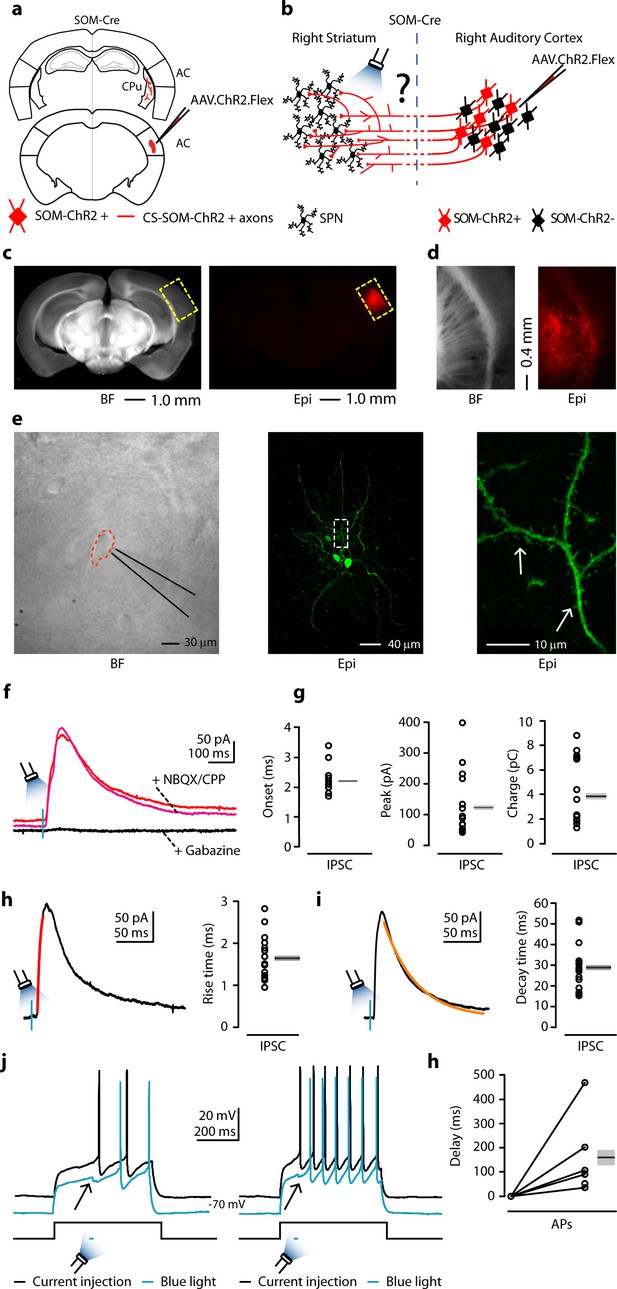
Photostimulation of auditory CS-SOM projections elicits direct inhibition and modulates action potentials in striatal SPNs.
(a) Schematic depicting injection site using the SOM-Cre transgenic mouse line to transfect CS-SOM projections to the dorsal striatum with ChR2. Bottom, auditory cortex: AAV.ChR2.flex injection site. Top, dorsal striatum: red CS-SOM ChR2-tdTomato-positive axons. (b) Experimental paradigm for photostimulating ChR2-positive CS-SOM projections while recording from SPNs. (c) Bright-field (left) and epifluorescence (right) images of a slice containing the auditory cortex injection site for AAV.ChR2.Flex. (d) Bright-field (left) and epifluorescence (right) images of a slice containing the dorsal striatum showing expression of ChR2-tdTomato following injection of AAV.ChR2.Flex into the auditory cortex. (e) Left, bright-field image of neurons as seen in bright-field microscopy during patch recordings. Middle, high-resolution epifluorescence image of a biocytin-labeled SPN. The dashed box indicates the location of the image in the right panel. Right, high-resolution epifluorescence image of spines from the biocytin-labeled SPN. (f) Example of IPSCs recorded at 0 mV from an SPN before (red trace) and after application of ionotropic glutamate receptor antagonists (NBQX 10 μM, CPP 5 μM: magenta trace) and GABAA receptor antagonist (gabazine 25 μM: black trace). (g) Left, plot of onset latencies recorded in SPNs (n = 16) including group averages (± s.e.m.). Middle, plot of IPSCs peaks calculated for SPNs, including group averages (± s.e.m.). Right, plot of IPSCs charge transfer calculated for individual IPSCs for SPNs, including group averages (± s.e.m.). (h) Left, example of IPSCs (black trace) and rising time course (red trace) recorded at 0 mV from an SPN. Right, plot of IPSCs rising time course recorded in SPNs (n = 16) including group averages (± s.e.m.). (i) Left, example of IPSCs (black trace) and decay time course (amber trace) recorded at 0 mV from an SPN. Right, plot of IPSCs decay time course recorded in SPNs (n = 16) including group averages (± s.e.m.). (j) Left (black trace), response of an SPN in the whole-cell current-clamp configuration to current injection (250 pA, 500 ms; n = 6). Left (blue trace), response of the SPN to current injection with photostimulation of CS-SOM projections (blue bar, 5–20 ms). Right (black trace), response of an SPN in the whole-cell current-clamp configuration to current injection (350 pA, 500 ms; n = 6). Left (blue trace), response of the SPN to current injection with photostimulation of CS-SOM projections (blue bar, 5–20 ms). (k) Summary of ChR2-mediated delay of action potential generation in SPNs (n = 6) during current injection combined with photostimulation of the ChR2 CS-SOM projections. Delay was relative to the onset of the first action potential measured during the current injection alone.
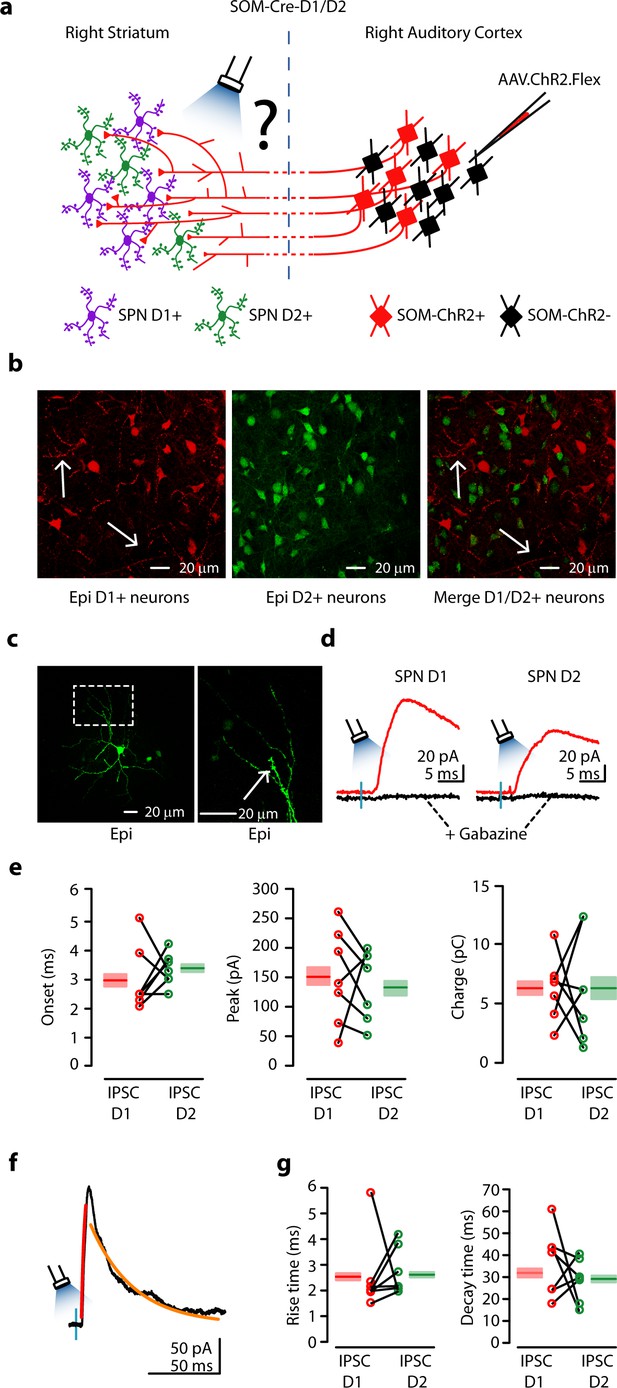
Auditory CS-SOM neurons innervate both dSPNs and iSPNs.
(a) Experimental paradigm for photostimulating ChR2-positive CS-SOM projections while recording from genetically labeled dSPNs and iSPNs. (b) Left, epifluorescence image of dSPNs expressing D1-tdTomato (white arrows indicate CS-SOM ChR2-tdTomato-positive axons). Middle, epifluorescence image of iSPNs expressing D2-EGFP. Right, overlay of D1-tdTomato (dSPNs) and D2-GFP (iSPNs). Note that the two sub-types of SPNs have a different distribution with no overlap when located in the same region of the dorsal striatum (white arrows indicate CS-SOM ChR2-tdTomato-positive axons). (c) Left, high-resolution epifluorescence image of a biocytin-labeled dSPN. The dashed box indicates the location of the image in right panel. Right, high-resolution epifluorescence image of spines from the biocytin-labeled dSPN. (d) Example of IPSCs recorded at 0 mV from a dSPN (left) and iSPN (right) after application of ionotropic glutamate receptor antagonists (NBQX 10 μM, CPP 5 μM: red traces) and GABAA receptor antagonist (gabazine 25 μM: black traces). (e) Left, plot of IPSCs onset latencies recorded in dSPNs (n = 7; red circles) and iSPNs (n = 7; green circles), including group averages (± s.e.m.). Middle, plot of IPSCs peaks calculated for dSPNs (n = 7; red circles) and iSPNs (n = 7; green circles), including group averages (± s.e.m.). Right, plot of IPSCs charge calculated for dSPNs (n = 7; red circles) and iSPNs (n = 7; green circles), including group averages (± s.e.m.). (f) Example of IPSCs (black trace), rising time course (red trace) and decay time course (amber trace) recorded at 0 mV from a dSPN. (g) Left, plot of IPSCs rising time course calculated for dSPNs (n = 7; red circles) and iSPNs (n = 7; green circles), including group averages (± s.e.m.). Right, plot of IPSCs decay time course calculated for dSPNs (n = 7; red circles) and iSPNs (n = 7; green circles), including group averages (± s.e.m.).
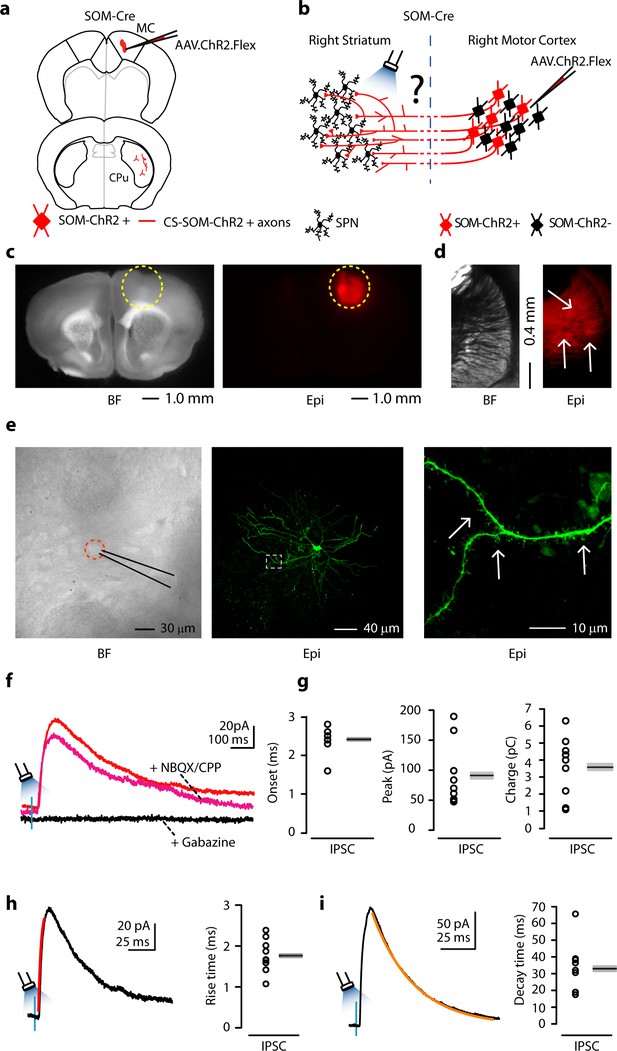
Photostimulation of motor CS-SOM projections elicits direct inhibition of striatal SPNs.
(a) Schematic depicting injection site using the SOM-Cre transgenic mouse line to transfect CS-SOM projections to the dorsal striatum with ChR2. Top, motor cortex: AAV.ChR2.flex injection site. Bottom, dorsal striatum: red CS-SOM ChR2-tdTomato-positive axons. (b) Experimental paradigm for photostimulating ChR2-positive CS-SOM projections while recording from SPNs. (c) Bright-field (left) and epifluorescence (right) images of a slice containing the motor cortex injection site for AAV.ChR2.Flex. (d) Bright-field (left) and epifluorescence (right) images of a slice containing the dorsal striatum showing expression of ChR2-tdTomato following injection of AAV.ChR2.Flex into the motor cortex. (e) Left, bright-field image of neurons as seen in bright-field microscopy during patch recordings. Middle, high-resolution epifluorescence image of a biocytin-labeled SPN. The dashed box indicates the location of the image in the right panel. Right, high-resolution epifluorescence image of spines from the biocytin-labeled SPN. (f) Example of IPSCs recorded at 0 mV from an SPN before (red trace) and after application of ionotropic glutamate receptor antagonists (NBQX 10 μM, CPP 5 μM: magenta trace) and GABAA receptor antagonist (gabazine 25 μM: black trace). (g) Left, plot of onset latencies recorded in SPNs (n = 9) including group averages (± s.e.m.). Middle, plot of IPSCs peaks calculated for SPNs, including group averages (± s.e.m.). Right, plot of IPSCs charge transfer calculated for individual IPSCs for SPNs, including group averages (± s.e.m.). (h) Left, example of IPSCs (black trace) and rising time course (red trace) recorded at 0 mV from an SPN. Right, plot of IPSCs rising time course recorded in SPNs (n = 9) including group averages (± s.e.m.). (i) Left, example of IPSCs (black trace) and decay time course (amber trace) recorded at 0 mV from an SPN. Right, plot of IPSCs decay time course recorded in SPNs (n = 9) including group averages (± s.e.m.).
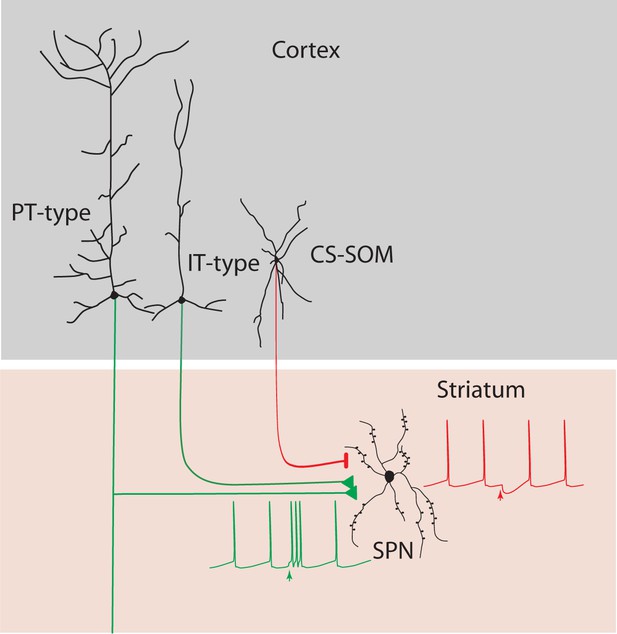
Summary diagram: CS-SOM neurons directly inhibit striatal SPNs.
Auditory and motor CS-SOM projections modulate the activity of striatal SPNs by direct inhibition. Green lines: excitatory inputs from intratelencephalic (IT-type) and projecting-type (PT-type) layer 5 pyramidal neurons; red line: inhibitory input from CS-SOM neurons.





