Inactivation of oncogenic cAMP-specific phosphodiesterase 4D by miR-139-5p in response to p53 activation
Figures
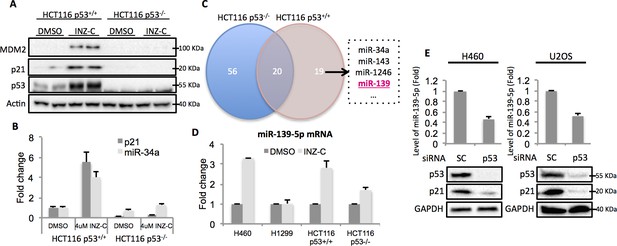
Identification of miR-139 as a novel p53-responsive gene.
Validation of induction of p53 and its targets gene. HCT116 p53+/+ and HCT116 p53-/- cells were treated with DMSO or 4uM INZ-C for 18 hr in duplicate and the protein samples and RNA samples were prepared for Western Blot (A) and qRT-PCR (B), respectively. (C). microRNA sequencing data analysis shows significantly induced microRNAs uniquely or commonly in HCT116 p53-/- and HCT116 p53+/+ cells. miR-139 was highlighted in red among other known p53 targets. (D). Induction of miR-139-5p by p53. p53 positive (H460, HCT116 p53+/+) and p53 negative (H1299, HCT116 p53-/-) cells were treated as in (A) and (B), and qRT-PCR analysis followed. (E). Decrease of miR-139-5p by knocking down p53. H460 and U2OS cells were transfected with scramble siRNA (SC) or siRNA specific to p53 (p53) and subjected to Western blot and qRT-PCR analysis.
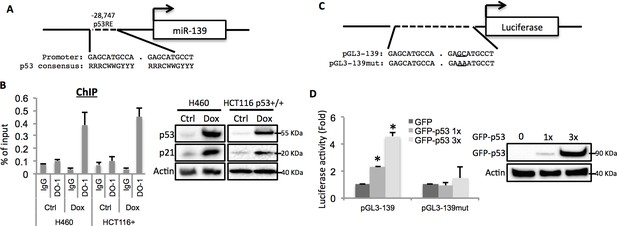
miR-139 is a direct target of p53.
(A) Diagram shows putative p53 responsive element (p53RE) located upstream of miR-139 gene. (B) Increased binding of p53 and the endogenous p53RE-containing miR-139 promoter in response to Doxorubicin. H460 or HCT116 p53+/+ cells were treated with 0.5μM Doxorubicin for 18 hr to stimulate the endogenous p53 before chromatin-associated immunoprecipitation assays were conducted with DO-1 p53 antibodies and the p53RE specific primers listed in Supplementary file 1 online. Western blot analysis was performed to confirm induction of p53. (C) Schematic of the pGL3 luciferase reporter constructs used. The plasmids either contain wild-type or mutant p53RE sequences of the miR-139 promoter, as underlined. (D). Enhancement of miR-139-promoter-driven luciferase activity by p53. H1299 cells were co-transfected with pGL3-139 or pGL3-139mut and increasing amount of GFP-p53 and collected 48 hr after transfection for assessment of luciferase activity, which was normalized against β-gal expression. Western blot was also performed to confirm the expression of GFP-p53. Error bars represent standard deviation (n = 3). β-gal, β-galactosidase; Ctrl, control; Dox, Doxorubicin; IgG, immunoglobulin G; mut, mutant.
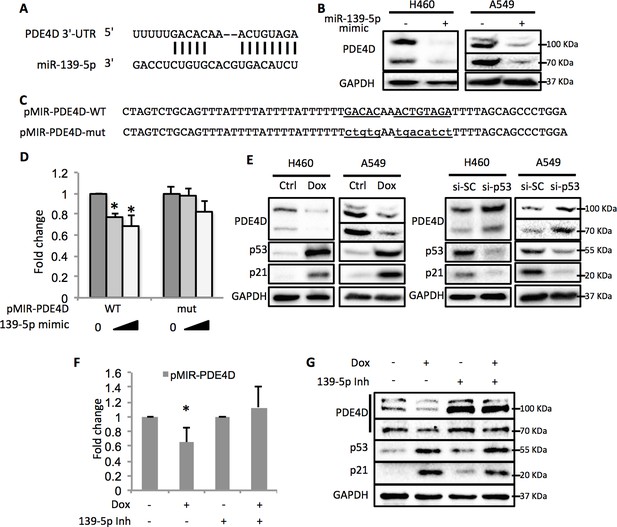
p53 modulates PDE4D expression via miR-139-5p.
(A) Bioinformatic analysis shows the miR-139-5p-targeted 3’-untranslated region (3’UTR) sequence of PDE4D mRNA. (B) Overexpression of miR-139-5p decreases the level of PDE4D protein in cells. H460 and A549 cells were collected 48 hr after transfection with miR-139-5p mimic for Western blot analysis. (C) Schematic of the pMIR–PDE4D luciferase reporter constructs used, which contain either a wild-type or a mutant miR-139-5p target site derived from the PDE4D mRNA. (D) Overexpression of miR-139-5p specifically inhibits luciferase activity from the plasmid harboring a wild-type, but not a mutant, miR-139-5p targeted sequence. H1299 cells were co-transfected with the indicated plasmids and collected 48 hr after transfection for luciferase assay. Luciferase activity was measured and normalized against β-gal expression. Even amount of oligos was achieved by adding Negative control oligos accordingly. (E) p53 modulates PDE4D expression. H460 and A549 cells were collected for Western blot analysis 18 hr after treatment with Doxorubicin (left panel) or 48 after transfection with si-SC or si-p53 (right panel). (F and G) p53 modulation of PDE4D expression is through miR-139-5p. H460 cells were co-transfected with indicated plasmids and 24 hr later were treated with Doxorubicin for 18 hr followed by measurement of luciferase activity (F) or Western blot analysis (G). 139-5p m, miR-139-5p mimic; 139-5p Inh, miR-139-5p inhibitor. Cells were treated with solvent for Dox or transfected with the same concentration of negative control oligos as miR-139-5p mimic or inhibitor. Error bars represent standard deviation (n = 3). *p<0.05.
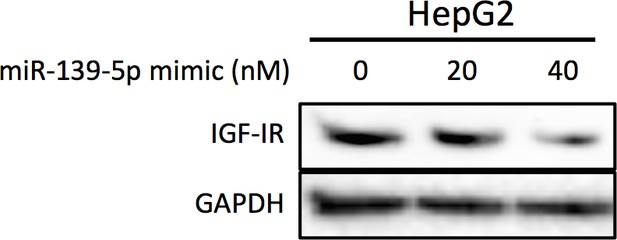
HepG2 cells were collected 48 hr after transfection with miR-139-5p mimic at 0, 20 or 40 nM for Western blot analysis on IGF-IR expression.
https://doi.org/10.7554/eLife.15978.006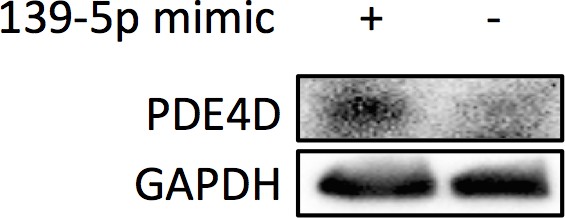
PC-3 cells were collected 48 hr after transfection with miR-139-5p mimic at 40 nM for Western blot analysis on PDE4D expression.
https://doi.org/10.7554/eLife.15978.007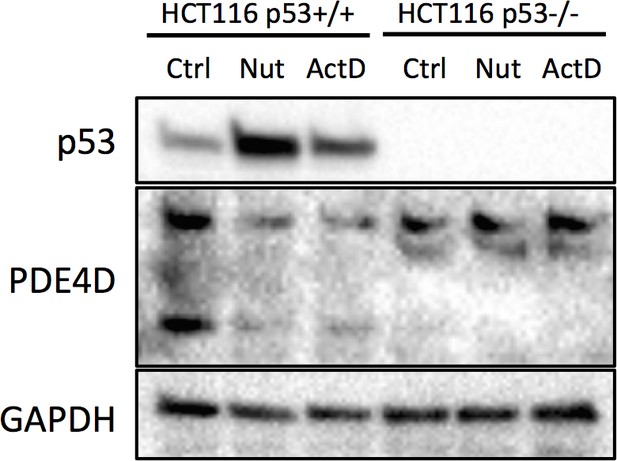
HCT116 p53+/+ and HCT116 p53-/- cells were treated with 10 μM Nutlin-3 (Nut) or 5 nM Actinomycin D (ActD) for 18 hr, and then collected for Western blot analysis.
https://doi.org/10.7554/eLife.15978.008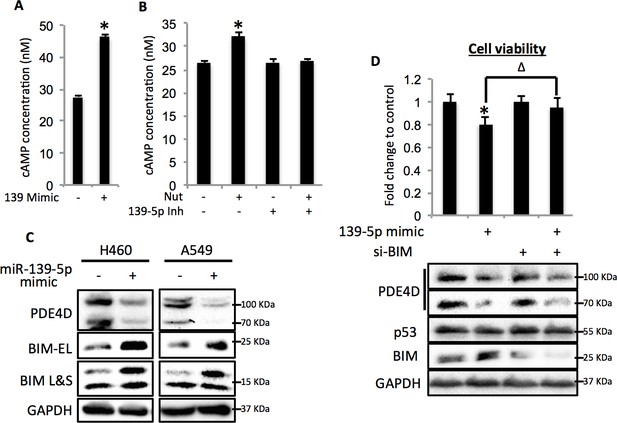
miR-139-5p induces cAMP/BIM mediated cell growth arrest.
(A). Overexpression of miR-139-5p increases cellular cAMP levels. A549 cells were transfected with miR-139-5p mimic control or miR-139-5p mimic and were subjected to cAMP measurement 48 hr after transfection. (B). Nutlin-3 increases cellular cAMP levels via miR-139-5p. A549 cells were transfected with miR-139-5p inhibitor control or miR-139-5p inhibitor for 48 hr followed by 10 μM Nutlin-3 treatment for 18 hr and subjected to cAMP measurement. (C). Overexpression of miR-139-5p induces BIM expression. H460 and A549 cells were transfected with miR-139-5p mimic and Western blot performed 48 hr after transfection. (D). Knockdown of BIM rescues miR-139-5p induced cell growth arrest. A549 cells were transfected with miR-139-5p mimic or siRNA against BIM (si-BIM) or both and subjected to MTT assay 48 hr after transfection. Cells were treated with solvent for Nutlin-3 or transfected with the same concentration of negative control oligos as miR-139-5p mimic or BIM siRNA. Error bars represent standard deviation (n = 3). *p<0.05 as compared to negative control oligos. Δ, p<0.05.
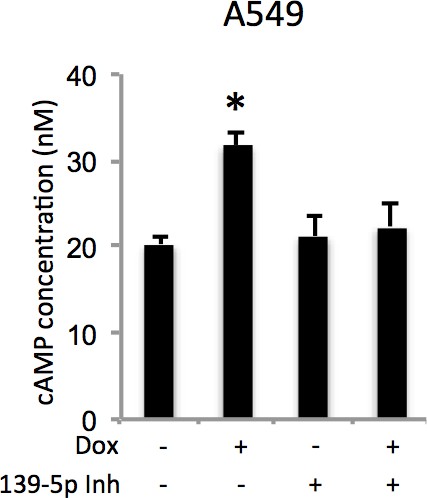
Doxorubicin increases cellular cAMP levels via miR-139-5p.
A549 cells were transfected with miR-139-5p inhibitor control or miR-139-5p inhibitor for 48 hr followed by 0.5 μM Doxorubicin treatment for 18 hr and subjected to cAMP measurement.
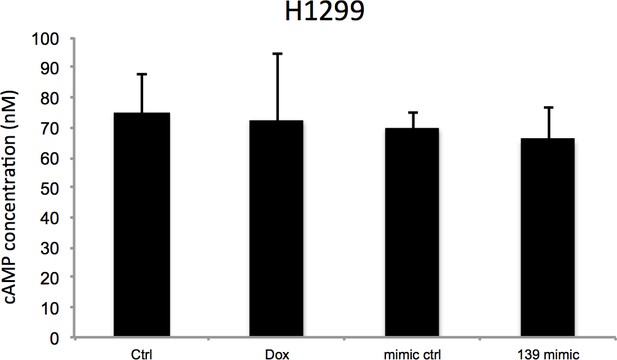
H1299 cells were treated with vehicle or 0.5 μM Doxorubicin for 18 hr followed by cAMP measurement, or transfected with miR-139-5p mimic control or miR-139-5p mimic and subjected to cAMP measurement 48 hr after transfection.
https://doi.org/10.7554/eLife.15978.011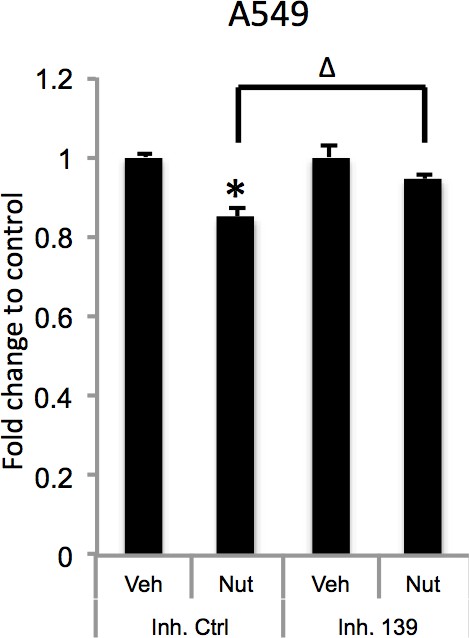
miR-139-5p inhibitor rescues Nutlin-3 inhibition of cell growth.
A549 cells were transfected with miR-139-5p inhibitor control or miR-139-5p inhibitor for 48 hr followed by 10 μM Nutlin-3 treatment for 24 hr and subjected to MTT analysis. *p<0.05 as compared to control siRNA. Δ, p<0.05.
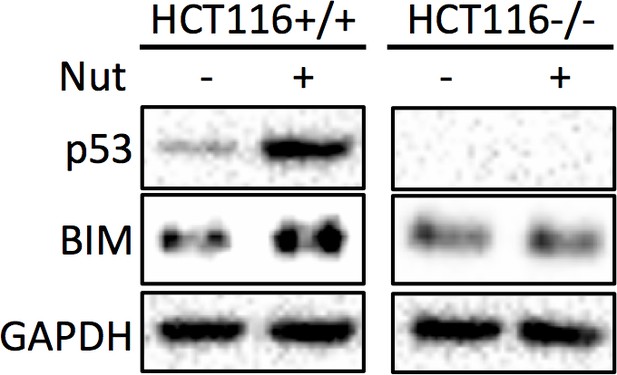
HCT116 p53+/+ and HCT116 p53-/- cells were treated with vehicle, or 10 μM Nutlin-3 for 18 hr, and subjected to Western blot analysis.
https://doi.org/10.7554/eLife.15978.013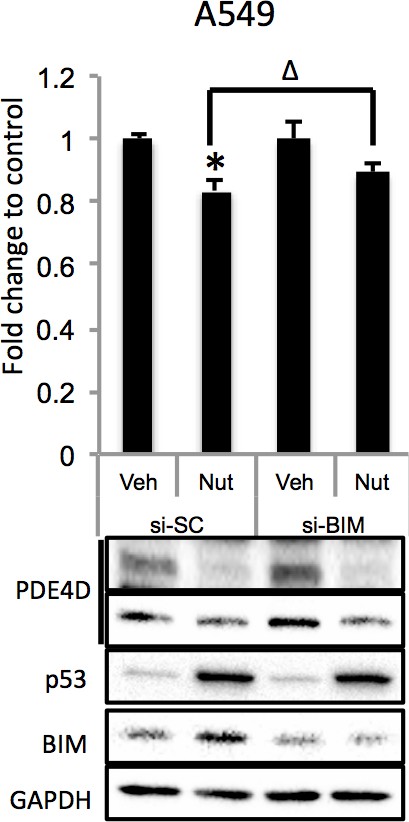
BIM knockdown alleviates Nutlin-3 inhibition of cell growth.
A549 cells were transfected with control siRNA or BIM siRNA and 48 hr after transfection, the cells were treated with 10 μM Nutlin-3 for 24 hr, followed by MTT assay and Western blot analysis. Error bars represent standard deviation (n = 3). *p<0.05 as compared to control siRNA. Δ, p<0.05.
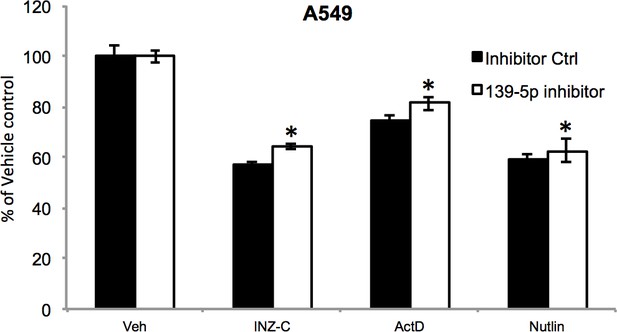
A549 cells were transfected with miRNA inhibitor control or miR-139-5p inhibitor, and 48 hr after transfection, the cells were treated with vehicle, 2 μM INZ-C, 5 nM ActD or 10 μM Nutlin-3 for 48 hr, followed by MTT assay to determine cell viability.
*p<0.05 as compared to the respective miRNA inhibitor control.
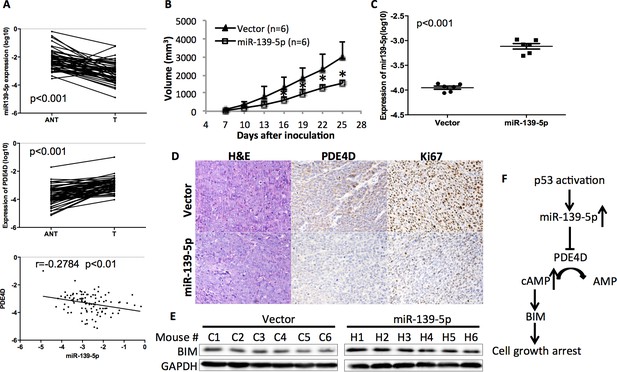
miR-139-5p is negatively correlated with PDE4D expression in human colorectal tumor samples, and represses the growth of SW480 xenograft tumors.
(A) miR-139-5p is negatively correlated with PDE4D expression in colon tumor samples. miR-139-5p (top panel) and PDE4D (middle panel) RNA expression was determined in 50 tumors (T) and paired adjacent normal tissues (ANT). The Pearson’s correlation of miR-139-5p and PDE4D RNA expression was analyzed combining tumor samples and normal tissues (bottom panel). (B) The sizes of SW480 xenograft tumors stably overexpressing pEZX-control (Vector) or pEZX-miR-139-5p (miR-139-5p) were measured every three days starting at seven days after inoculation. Mean tumor sizes were presented. Error bar, SD; *p<0.05. (C) Expression of miR-139-5p of xenograft tumors was determined by qRT-PCR. (D) Representative images of the xenograft tumors stained with hematoxylin and eosin (H&E), or immunohistochemistry analyzed with PDE4D or Ki67 antibody. (E) Xenograft tumors were subjected to Western blot analysis of BIM expression. (F) Proposed model of the p53/miR-139-5p/PDE4D pathway.
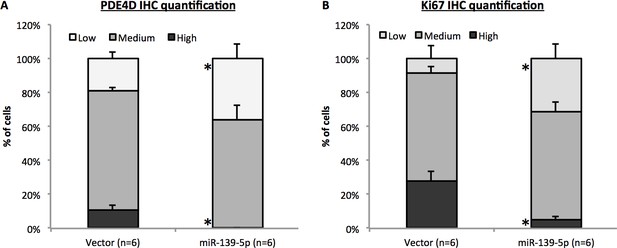
Quantification of PDE4D and Ki67 IHC analysis.
The IHC staining intensity was categorized to low, medium and high as determined by ImageJ software. Five random fields were chosen from each slide to obtain average intensity of each category, and all six mice from each group were included in the analysis. *p<0.05 as compared to control.
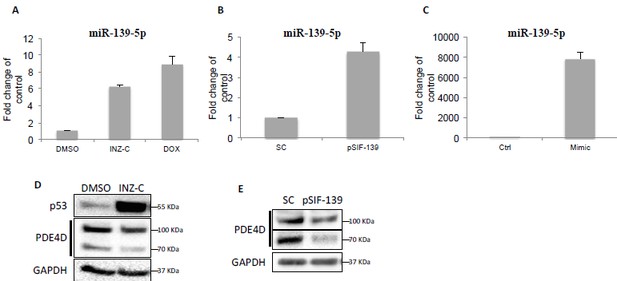
Downregulation of PDE4D by p53 and miR-139-5p.
(A) H460 cells were treated with DMSO, 2 μM INZ-C or 0.5 μM Dox for 18 hr followed by qRT-PCR analysis of miR-139-5p. (B) and (C) H460 cells were transfected with pSIF-H1-Scramble control (SC) or pSIF-H1-miR-139-5p (pSIF-139) (B), or negative control oligos (Ctrl) or miR-139-5p mimic (Mimic), followed by qRT-PCR analysis of miR-139-5p at 48 hr after transfection. Samples from (A) and (B) were also analyzed by Western blot as shown in (D) and (E), respectively.
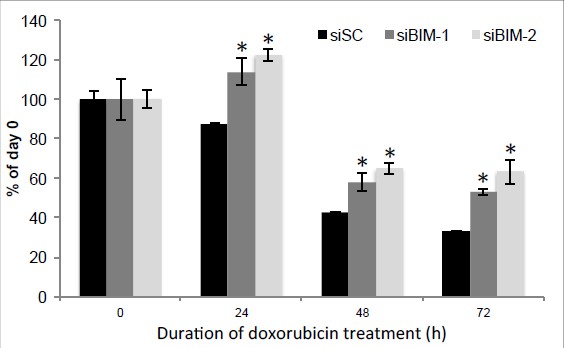
Knockdown of BIM rescue doxorubicin suppression of H460 cell proliferation.
H460 cells were transfected with two siRNAs against BIM, and 48 hr after transfection, cells were treated with 0.5 μM Dox for indicated duration and cell proliferation was determined by using the sulforhodamine B (SRB) assay. * p<0.05. Error bars, SD.
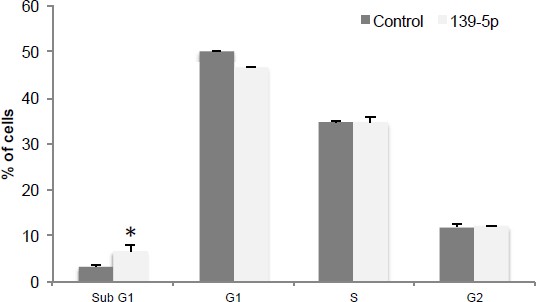
miR-139-5p induces apoptosis.
H460 cells were transfected with pSIF-H1-Scramble control (Control) or pSIF-H1-miR-139-5p (139-5p), and 48 hr after transfection, the cells were subjected to flow cytometry analysis. *p<0.05. Error bars, SD.
Additional files
-
Supplementary file 1
List of primers used in this study.
- https://doi.org/10.7554/eLife.15978.018





