The composition and organization of Drosophila heterochromatin are heterogeneous and dynamic
Figures
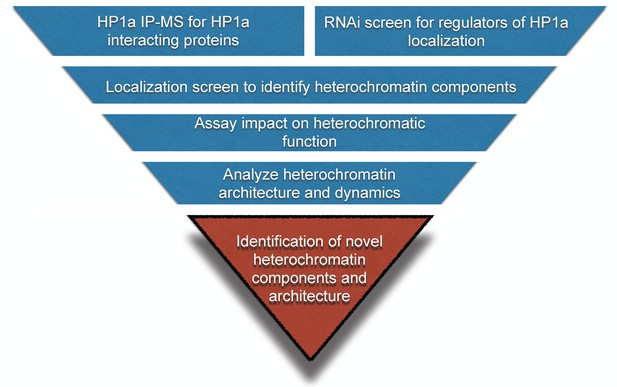
Workflow to identify novel heterochromatin components and regulators.
We devised an unbiased strategy to identify novel components of heterochromatin. First, we identified candidates by performing HP1a immunoprecipitation followed by mass spectrometry (IP-MS) and a genome-wide RNAi screen. Candidates that localized to heterochromatin were assayed for effects on PEV. Finally, we investigated their spatial and temporal localization with respect to heterochromatin.
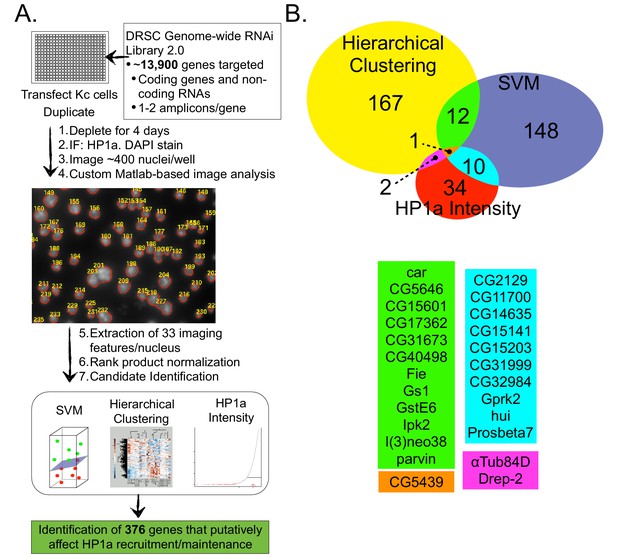
A genome-wide image-based RNAi screen identifies HP1a regulators.
Drosophila Kc cells transfected with dsRNA were analyzed for HP1a localization by IF, and DNA was counterstained with DAPI. Cells were visualized using high-throughput fluorescent microscopy and imaging features were extracted using custom Matlab scripts. Wells were normalized and checked for replicate consistency using the Rank Product test and a p-value was calculated. Putative candidates involved in HP1a recruitment/maintenance were selected by identifying amplicons that lowered HP1a intensity, or clustered with HP1a depletions after hierarchical clustering or Support Vector Machine (SVM) analysis. (B) Genes that clustered using unsupervised hierarchical clustering with either HP1a or Su(var)3–9 positive control depletions are represented by the yellow circle. Supervised machine learning models (SVMs) were trained to identify genes that disrupt HP1a staining (blue circle) using HP1a depletion controls. HP1a intensity measures (mean, maximum, relative maximum and kurtosis) were used to identify another set of candidate genes (red circle). Genes identified by multiple methods are indicated by color below the Venn diagram. See Figure 2—source data 1 for a list of all genes identified in the RNAi screen and the method used to identify them.
-
Figure 2—source data 1
374 genes putatively regulate heterochromatin.
Genes identified from the RNAi screen whose depletion results in reduced HP1a levels or phenocopies HP1a or Su(var)3–9 depletions (HP1a positive regulators, HPprs) and the method used to identify them are listed.
- https://doi.org/10.7554/eLife.16096.009
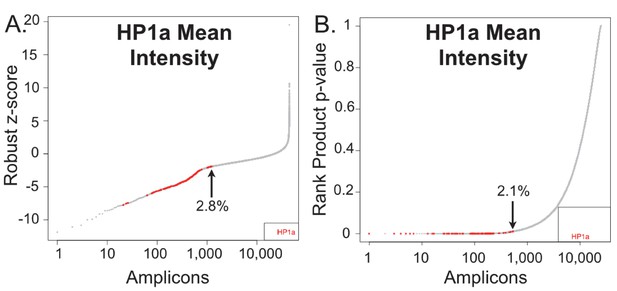
The rank product test is more effective than the robust Z-Score at identifying HP1a knockdowns.
HP1a mean intensity was normalized using the robust z-score (A) or the rank product test (B). The normalized value (or p-value) is plotted versus a ranked list of the amplicons, with a value of one indicating the strongest hit. HP1a RNAi (positive controls) are noted in red and the percentage for the highest ranked positive control is indicated with an arrow.
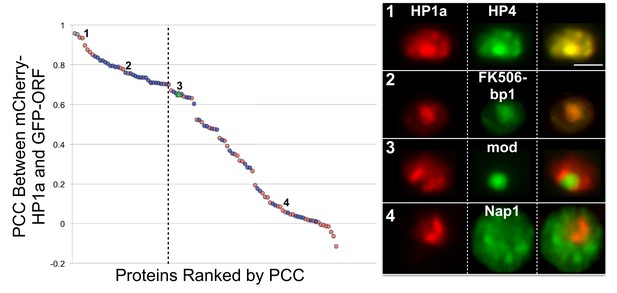
Identification of candidates that co-localize with HP1a.
Proteins were selected from the HP1a IP-MS (red circles) or the RNAi screen (blue circles), tagged with GFP (green), and analyzed for localization with respect to mCherry-tagged HP1a (red). GFP-tagged HP1a was used as a positive control (gray circles). The Pearson correlation coefficient (PCC) between mCherry-HP1a and GFP-tagged proteins left of the dashed line was significantly higher than the PCC between mCherry-HP1a and GFP-mod (green triangle), using the two-sided unpaired Mann-Whitney test (p-value<0.05). Numbers on graph correspond to representative images (right panel). Scale bar is 5 µm. See Figure 3—source data 1 for the PCC of all proteins tested.
-
Figure 3—source data 1
Identification of candidates that co-localize with HP1a.
Proteins were selected from the HP1a IP-MS or the RNAi screen, tagged with GFP and analyzed for localization with respect to mCherry-tagged HP1a. GFP-tagged HP1a was used as a positive control. Cells shaded red were found to have a significant increase in the Pearson correlation coefficient (PCC) between GFP-mod (dark green shading) and GFP-ORF PCC using the two-sided unpaired Mann-Whitney test (p-value<0.05).
- https://doi.org/10.7554/eLife.16096.013
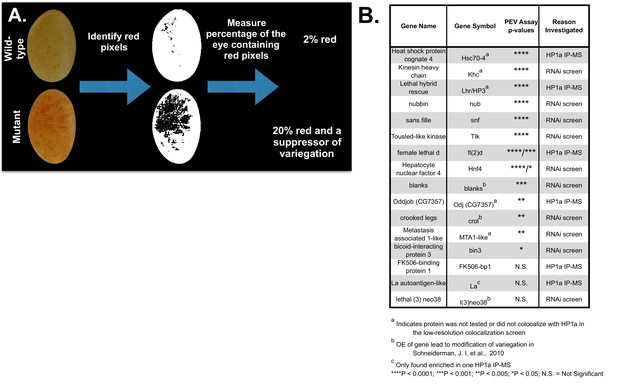
HPips and RNAi screen candidates are suppressors of variegation.
(A) Color Inspector 3D in ImageJ was used to determine the RGB values of 'red' pixels (indicating loss of suppression). The percent of the eye composed of red pixels was then calculated. (B) Fly mutants and RNAi lines were tested for impact on white variegation in y, w, KV108 males, and are organized by p-value. Mutations were tested for dominant effects if they were recessive lethal, otherwise homozygotes were analyzed. CG7357[f00521] was scored for variegation using the yellow reporter gene, since the line harbors a mini-white reporter that precludes assessment of white variegation. The p-values were calculated using a 2-tailed, 2-sample unequal variance t-test for white variegation and a 2-sample Kolmogorov-Smirnov test for yellow variegation. Positive and negative controls were performed and are listed in the Figure 4—source data 1 along with the genotypes of all the fly lines used. CG2129, Ssrp and Ref1 could not be tested for effects on variegation using RNAi lines, due to lethality.
-
Figure 4—source data 1
HPips and RNAi screen candidates are suppressors of variegation.
Fly mutants and RNAi lines were tested for white variegation in y, w, KV108 males and are grouped with the appropriate positive and negative control(s). Mutants were only tested for dominant effects if they were recessive lethal. See legend of Figure 4 for additional details.
- https://doi.org/10.7554/eLife.16096.015
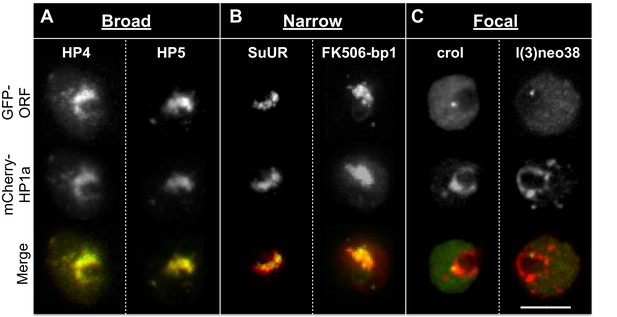
Heterochromatic proteins display diverse localization patterns.
HP4 and HP5 broadly overlap with HP1a. SuUR and FK506-bp1 overlap with the interior of HP1a (narrow). Crol and l(3)neo38 form a focus within the HP1a domain (focal). Focal proteins are presented as slices, broad and narrow proteins are projections. mCherry-tagged HP1a is in red, GFP-tagged ORF is in green. Scale bar is 5 µm.
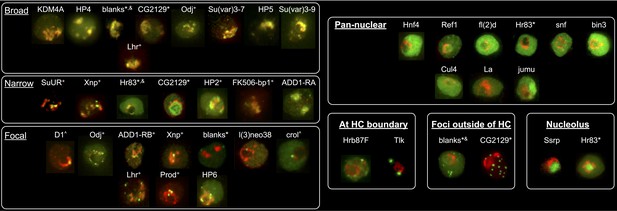
Heterochromatic proteins display diverse localization patterns.
mCherry-tagged HP1a is in red, GFP-tagged ORF is in green. Localization patterns are grouped into 7 categories: 'Broad' - almost complete overlap with HP1a; 'Narrow' - only partial overlap with HP1a; 'Focal' - forms a foci or focus that overlap with HP1a; 'Pan-nuclear' - everywhere in the nucleus; 'At HC boundary' - enriched at the periphery of heterochromatin; 'Foci outside of HC' - forms foci outside of heterochromatin; and 'Nucleolus' - localizes to the nucleolus. Proteins labelled with + indicates that in a population of S2 cells they display patterns that fit in more than one category. * indicates that the localization pattern is dependent on cell type or location (N- or C-term) of the tag. Proteins labelled with ^ indicate a slice, otherwise images are projections. Prod was tagged with mCherry and image was false-colored; red indicates GFP-tagged HP1a and green indicates mCherry-tagged prod. & indicates experiment done in Kc cells. Hr83: narrow when N-terminal tagged in Kc cells (stable), pan-nuclear when N-term tagged in S2 cells (transient), nucleolar when C-term tagged in S2 cells (transient); CG2129: N-terminally GFP tagged construct (transient Kc and S2) shows nucleolar localization in lowly expressing cells and foci appear in highly expressing cells; blanks when N-term tagged in stable Kc cells broadly co-localizes with HP1a, N-term tagged in transient S2 has foci next to the HP1a domain, C-term tagged in transient S2 or Kc cells is pan-nuclear, antibody staining is also pan-nuclear with some structure throughout the nucleus. Su(var)3–9 is tagged with mClover. BioTAP tagged ADD1-RA (red) was visualized using a Peroxidase antibody followed by immunofluorescence.
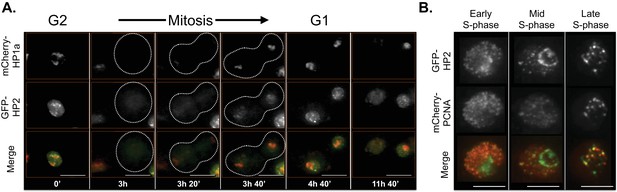
HP2 time-lapse imaging reveals dynamic regulation and overlap with PCNA throughout S-phase.
HP2 partially overlaps and is enriched at the boundary of HP1a in G2, released from chromatin during mitosis and broadly colocalized with HP1a during G1. Mitosis is used to discriminate G1 from G2. Dotted lines indicate the cell periphery during mitosis. mCherry-tagged HP1a is in red, GFP-tagged HP2 is in green. Scale bar is 10 µm. (B) HP2 overlaps with PCNA foci in early, mid and late S-phase. Representative images of early, mid and late S-phase are shown. mCherry-tagged PCNA is in red, GFP-tagged HP2 is in green. Scale bar is 5 µm.
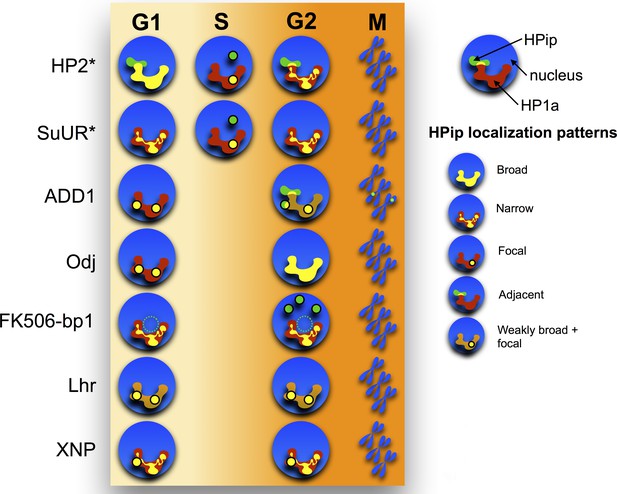
Time-lapse imaging reveals a variety of dynamic localization patterns within heterochromatin.
A graphical representation of the localization patterns of heterochromatic proteins throughout the cell cycle is shown. HP1a is depicted in red, the heterochromatin protein (HPip) in green and overlap between the two in yellow. A dotted circle indicates that FK506-bp1 forms a ring around the nucleolus. * indicates foci overlap completely with PCNA during S-phase.
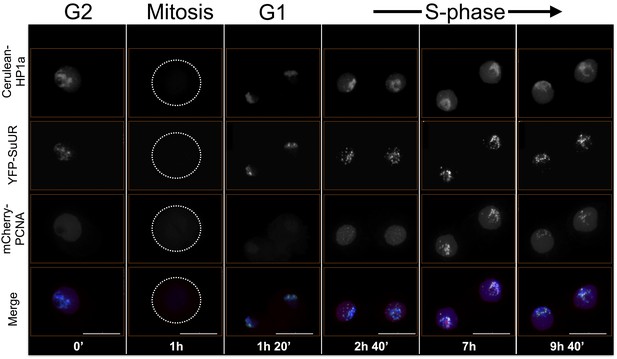
Combined SuUR, HP1a and PCNA time-lapse imaging reveals dynamic regulation.
SuUR colocalizes with HP1a during G2 and G1, and colocalizes with PCNA during S-phase. Dotted lines indicate the cell periphery during mitosis. Mitosis is used to discriminate G1 from G2, while PCNA foci indicates S-phase. Cerulean tagged HP1a is in blue, YFP tagged SuUR is in green, mCherry tagged PCNA is in red. Scale bar is 10 µm.
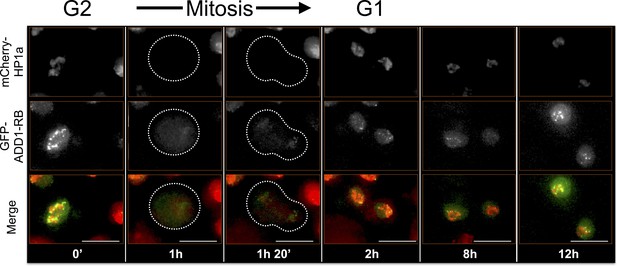
ADD1-PB time-lapse imaging reveals dynamic regulation.
ADD1-PB forms focal subdomains that abut and overlap HP1a, and does not overlap with the centromeric or telomeric markers CID and HOAP (data not shown), respectively. In G2 ADD1-PB is predominantly focal at the heterochromatin boundary. A small amount of discrete signal remains on chromatin during mitosis and persists at low levels into G1, before eventually increasing in intensity, which suggests loading at the end of G1 or during S-phase. Mitosis is used to discriminate G1 from G2. Dotted lines indicate the cell periphery during mitosis as it divides into two daughter cells (G1). mCherry tagged HP1a is in red, GFP tagged ADD1-PB is in green. Scale bar is 10 µm.
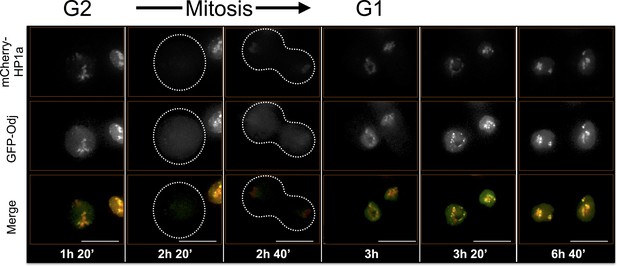
Oddjob time-lapse imaging reveals dynamic regulation.
Odj broadly co-localizes with HP1a at the end of G2 and disperses from chromosomes during mitosis. It reforms as a focal subdomain after mitosis that gradually increases in size, until it broadly overlaps HP1a again. Mitosis is used to discriminate G1 from G2. Dotted lines indicate the cell periphery during mitosis as it divides into two daughter cells (G1). mCherry tagged HP1a is in red, GFP tagged Odj is in green. Scale bar is 10 µm.
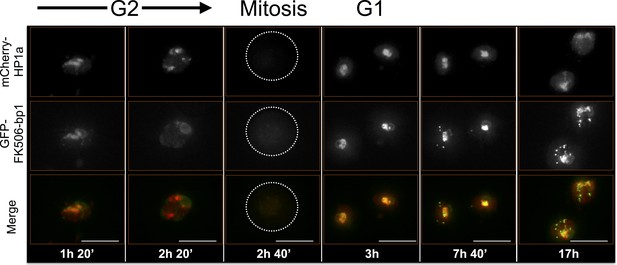
FK506-bp1 time-lapse imaging reveals dynamic regulation.
FK506-bp1 narrowly co-localizes with HP1a throughout much of the cell cycle and loses co-localization with HP1a 20 min to 1 hr before HP1a is released from chromosomes (prophase). After mitosis, the narrow co-localization pattern of FK506-bp1 is restored, with a weak ring around the nucleolus, which is located adjacent to the HP1a domain. FK506-bp1 foci then begin to accumulate outside of heterochromatin until just before prophase, when they disappear prior to HP1a removal. Foci do not track with PCNA (replication), CID (centromeres) or HOAP (telomeres) foci (data not shown). Mitosis is used to discriminate G1 from G2. Dotted lines indicate the cell periphery during mitosis as it divides into two daughter cells (G1). mCherry tagged HP1a is in red, GFP tagged FK506-bp1 is in green. Scale bar is 10 µm.
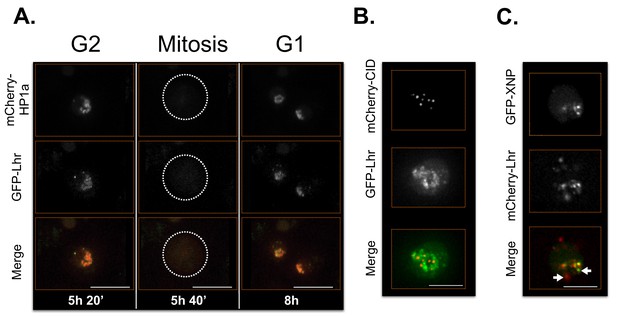
Lhr time-lapse imaging reveals dynamic regulation.
Lhr broadly co-localizes with HP1a and is released from chromatin during mitosis. Mitosis is used to discriminate G1 from G2. Dotted lines indicate the cell periphery during mitosis as it divides into two daughter cells (G1). mCherry tagged HP1a is in red, GFP tagged Lhr is in green. Scale bar is 10 µm. (B) Lhr partially overlaps centromeres. mCherry tagged CID is in red, GFP tagged Lhr is in green. Scale bar is 5 µm. (C) Some Lhr and XNP foci overlap, but others do not (arrows). mCherry tagged Lhr is in red, GFP tagged XNP is in green. Scale bar is 5 µm.
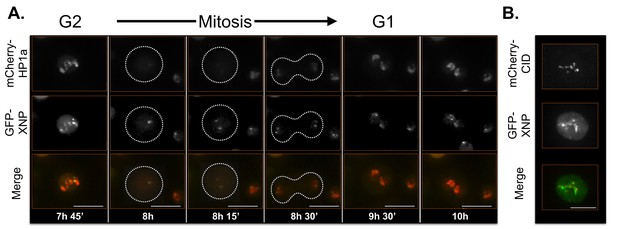
XNP time-lapse imaging reveals dynamic regulation.
XNP colocalizes with a portion of HP1a in G2. The majority of XNP is removed during mitosis, however 1–2 foci remain chromatin-bound. In G1 XNP is focal within the HP1a domain while gradually accumulating and colocalizing with more HP1a. Mitosis is used to discriminate G1 from G2. Dotted lines indicate the cell periphery during mitosis as it divides into two daughter cells (G1). mCherry-tagged HP1a is in red, GFP-tagged XNP is in green. Scale bar is 10 µm. (B) XNP and CID partially overlap. mCherry-tagged CID is in red, GFP-tagged XNP is in green. Scale bar is 5 µm.
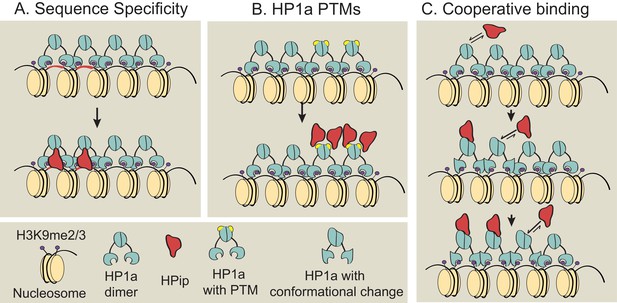
Models for subdomain formation within heterochromatin.
We propose three non-mutually exclusive models for subdomain formation of HP1a interacting proteins (HPips) within the HP1a (teal) heterochromatin holodomain. (A) The HPip (red) may be recruited to a specific sequence and seeds the formation of a subdomain (as observed for D1 [Aulner et al., 2002] and GAGA [Raff et al., 1994] factor). (B) HP1a and its orthologs are extensively post-translationally modified by SUMOylation, acetylation, methylation, formylation, ubiquitination and poly(ADP-ribosyl)ation (Alekseyenko et al., 2014; Lomberk et al., 2006; LeRoy et al., 2009). An HPip could have an increased binding affinity for a specific HP1a PTM (yellow). Thus, HP1a PTMs may regulate HP1a complex formation and spatially restrict HPip recruitment. Consistent with the PTM model, HP2 and PIWI have been shown to have higher binding affinities for HP1a proteins containing phospho-mimic mutations in the HP1a chromo shadow domain (Mendez et al., 2011). (C) Subdomains could form by a cooperative binding mechanism (Bray and Duke, 2004; Bai et al., 2010). HP1a can oligomerize at least up to tetramers (Wang et al., 2000; Zhao et al., 2000; Canzio et al., 2011), forming a multivalent platform for HPip binding (i.e. more than one HPip binding site per HP1a oligomer). Thus, initial binding by an HPip could induce a higher binding affinity between a neighboring HP1a molecule and the HPip. The dotted arrow indicates potential self-interactions between HPips and solid arrows indicate hypothetical HPip on/off rates.
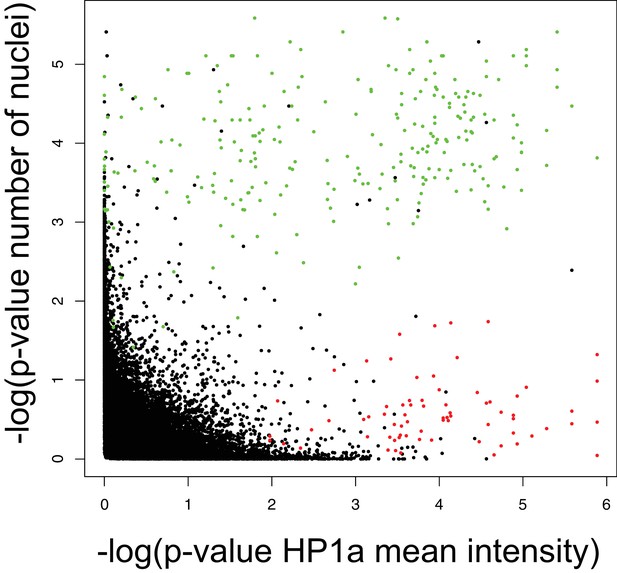
HP1a staining is significantly decreased in dying cells.
The negative log of the RP p-value for HP1a mean intensity and number of nuclei were plotted against each other. Green dots indicate thread depletions (dying cells) and red dots indicate HP1a depletions.
Videos
HP2 time-lapse imaging reveals dynamic regulation throughout the cell cycle.
HP2 partially overlaps and is enriched at the boundary of HP1a in G2, released from chromatin during mitosis and broadly colocalized with HP1a during G1. Mitosis is used to discriminate G1 from G2. mCherry-tagged HP1a is in red, GFP-tagged HP2 is in green. Scale bar is 10 µm.
Combined SuUR, HP1a and PCNA time-lapse imaging reveals dynamic regulation.
SuUR colocalizes with HP1a during G2 and G1, and colocalizes with PCNA during S-phase. Mitosis is used to discriminate G1 from G2, while PCNA foci indicate S-phase. Cerulean-tagged HP1a is in blue, YFP-tagged SuUR is in green, mCherry-tagged PCNA is in red. Scale bar is 5 µm.
ADD1-PB time-lapse imaging reveals dynamic regulation.
ADD1-PB forms focal subdomains that abut and overlap HP1a, b not overlap with the centromeric or telomeric markers CID and HOAP (data not shown), respectively. In G2 ADD1-PB is predominantly focal at the heterochromatin boundary. A small amount of discrete signal remains on chromatin during mitosis and persists at low levels into G1, before eventually increasing in intensity, which suggests loading at the end of G1 or during S-phase. Mitosis is used to discriminate G1 from G2. mCherry-tagged HP1a is in red, GFP-tagged ADD1-PB is in green. Scale bar is 5 µm.
Oddjob time-lapse imaging reveals dynamic regulation.
Odj broadly co-localizes with HP1a at the end of G2 and disperses from chromosomes during mitosis. It reforms as a focal subdomain after mitosis that gradually increases in size, until it broadly overlaps HP1a again. Mitosis is used to discriminate G1 from G2. mCherry-tagged HP1a is in red, GFP-tagged Odj is in green. Scale bar is 5 µm.
FK506-bp1 time-lapse imaging reveals dynamic regulation.
FK506-bp1 narrowly co-localizes with HP1a throughout much of the cell cycle and loses co-localization with HP1a 20 min to 1 hr before HP1a is released from chromosomes (prophase). After mitosis, the narrow co-localization pattern of FK506-bp1 is restored, with a weak ring around the nucleolus, which is located adjacent to the HP1a domain. FK506-bp1 foci then begin to accumulate outside of heterochromatin until just before prophase, when they disappear prior to HP1a removal. Foci do not track with PCNA (replication), CID (centromeres) or HOAP (telomeres) foci (data not shown). Mitosis is used to discriminate G1 from G2. mCherry-tagged HP1a is in red, GFP-tagged FK506-bp1 is in green. Scale bar is 10 µm.
Lhr time-lapse imaging reveals dynamic regulation.
Lhr broadly co-localizes with HP1a and is released from chromatin during mitosis. Mitosis is used to discriminate G1 from G2. mCherry-tagged HP1a is in red, GFP-tagged Lhr is in green. Scale bar is 5 µm.
XNP time-lapse imaging reveals dynamic regulation.
XNP colocalizes with a portion of HP1a in G2. The majority of XNP is removed during mitosis, however 1-2 foci remain chromatin-bound. In G1 XNP is focal within the HP1a domain but gradually accumulates in size and colocalizes with more HP1a. Mitosis is used to discriminate G1 from G2. mCherry-tagged HP1a is in red, GFP-tagged XNP is in green. Scale bar is 5 µm.
Tables
HP1a interactors ranked by frequency of detection. The most common HP1a interacting proteins are listed according to the frequency in which they were detected in HP1a IP-MS experiments (out of six). References that link a protein to HP1a by IP, yeast-two-hybrid or immunofluorescence are listed in the third column. Asterisk indicates that the protein has been shown to modulate PEV. See Table 1—source data 1 and 2 for a complete list of hits and Table 1—source data 3 for a silver-stained gel of the IP.
| Flybase Gene Name | # of experiments enriched in | Literature Linking the Gene to HP1a |
|---|---|---|
| ADD1* | 6 | Alekseyenko et al., 2014 |
| CG8108 | 6 | Alekseyenko et al., 2014; Guruharsha et al., 2011 |
| HP5* | 6 | Greil et al., 2007; Alekseyenko et al., 2014 |
| Su(var)3-9* | 6 | Schotta et al., 2002; Alekseyenko et al., 2014 |
| Su(var)2-HP2* | 6 | Shaffer et al., 2002; Alekseyenko et al., 2014 |
| tsr | 6 | |
| Hsc70-4 | 5 | Alekseyenko et al., 2014 |
| Kdm4A* | 5 | Lin et al., 2008; Alekseyenko et al., 2014; Colmenares et al., unpublished |
| Odj (CG7357) | 5 | van Bemmel et al., 2013 |
| smt3 | 4 | Alekseyenko et al., 2014 |
| Lhr | 4 | Greil et al., 2007; Alekseyenko et al., 2014 |
| Act5C | 4 | |
| Hsc70-3 | 4 | |
| betaTub56D | 4 | |
| Chd64 | 4 | |
| Hsp83 | 4 | |
| CG7692 | 3 | Alekseyenko et al., 2014 |
| HP4* | 3 | Greil et al., 2007 |
| Tudor-SN | 3 | |
| His2B:CG33872 | 3 | |
| eIF-4a | 3 | |
| FK506-bp1 | 3 | |
| CG7172 | 3 | |
| CG8258 | 3 | |
| EF2 | 3 | |
| eIF-4B | 3 | |
| Hsc70-5 | 3 | |
| Hsp60 | 3 | |
| qm | 3 | |
| sta | 3 |
-
Table 1—source data 1
2-Step HP1a IP-MS.
HP1a was purified in the absence of ionizing radiation (IR) (A), and 10 min (B) and 60 (C) minutes after 10 Gy exposure. Number of unique peptides per protein are listed. The HPips identified did not change significantly with respect to irradiation, therefore we used all purifications to identify candidate hits.
- https://doi.org/10.7554/eLife.16096.005
-
Table 1—source data 2
1-Step HP1a IP-MS.
HP1a was purified in the absence of IR (A, Mock and FS-HP1a), and 10 (B, FS-HP1a) and 60 (C, FS-HP1a) minutes after 10 Gy exposure. Number of unique peptides per protein is listed. The HPips identified did not change significantly with respect to irradiation, therefore we used all purifications to identify candidate hits.
- https://doi.org/10.7554/eLife.16096.006
-
Table 1—source data 3
HP1a interacts with a large set of proteins.
Silver-stained gel of a single step purification from S2 cells stably expressing FS-HP1a (lanes 1–3) or WT (lane 4) S2 cells. HP1a was purified in the absence of IR (lane 1), and 10 (lane 2) and 60 (lane 3) minutes after 10 Gy exposure. The HPips identified did not change significantly with respect to irradiation, therefore all purifications were used to identify candidate hits.
- https://doi.org/10.7554/eLife.16096.007
RNAi screen hits with previously known connections to heterochromatin. Identified hits from the RNAi screen with previously known connections to heterochromatin are listed according to the method of identification (Hierarchical Clustering, HP1a Intensity or Support Vector Machine [SVM]). Whether a gene clustered with HP1a or Su(var)3–9 depletion controls after Hierarchical Clustering is indicated in parentheses.
| Flybase Name or Symbol | Method of Identification | Link to Heterochromatin | Reference |
|---|---|---|---|
| Ssrp | Hierarchical Clustering (HP1a, Su(var)3-9) | Part of FACT complex | Orphanides et al., 1999 |
| MBD-like | Hierarchical Clustering (HP1a) | Repressive, localizes to chromocenter, part of NuRD complex | Ballestar et al., 2001; Marhold et al., 2004 |
| stellate | Hierarchical Clustering (HP1a) | Subunit of Casein kinase II | Bozzetti et al., 1995 |
| kismet | Hierarchical Clustering (HP1a) | Su(var), regulates heterochromatic silencing | Schneiderman et al., 2009, 2010 |
| Spt20 | Hierarchical Clustering (HP1a) | Part of SAGA complex | Weake et al., 2009 |
| Su(var)205 | Hierarchical Clustering (HP1a), HP1a Intensity, SVM | Encodes HP1a | |
| l(3)neo38 | Hierarchical Clustering (HP1a), SVM | Regulates heterochromatic silencing | Schneiderman et al., 2010 |
| Hdac3 | Hierarchical Clustering (Su(var)3-9) | Ortholog regulates HP1beta levels | Bhaskara et al., 2010 |
| Rm62 (lip, p68) | Hierarchical Clustering (Su(var)3-9) | Su(var), binds and putatively targets Su(var)3-9, binds blanks, binds AGO2, regulates heterochromatic silencing | Csink et al., 1994; Boeke et al. 2011; Gerbasi et al., 2011; Ishizuka et al., 2002; Schneiderman et al., 2010 |
| jumu | Hierarchical Clustering (Su(var)3-9) | Localizes to chromocenter, modifier of variegation | Hofmann et al., 2010, 2009 |
| MTA1-like | Hierarchical Clustering (Su(var)3-9) | Part of NuRD complex | Marhold et al., 2004 |
| AGO2 | Hierarchical Clustering (Su(var)3-9) | Heterochromatin targeting, Su(var) | Noma et al., 2004; Deshpande et al., 2005 |
| moi | Hierarchical Clustering (Su(var)3-9) | Protects telomeres | Raffa et al., 2009 |
| Adar | HP1a Intensity | E(var) on the 4th chromosome, edits RNA, silences Hoppel's transposase | Savva et al., 2013 |
| Parp | HP1a Intensity | E(var), promotes chromatin condensation and represses retrotransposons | Tulin and Spradling, 2003 |
| Ino80 | HP1a Intensity | Ortholog in mice complexed with YY1 which regulates HP1gamma, regulates heterochromatic silencing | Wu et al., 2009; Schneiderman et al., 2010 |
| roX1 | HP1a Intensity | Su(var) | Deng et al., 2009 |
| modulo | SVM | Localizes to chromocenter, Su(var) | Perrin et al., 1998; Garzino et al., 1992 |
| blanks | SVM | Regulates heterochromatic silencing | Schneiderman et al., 2010 |
| crol | SVM | Regulates heterochromatic silencing | Schneiderman et al., 2010 |
| Samuel | SVM | Regulates heterochromatic silencing | Schneiderman et al., 2010 |
| Wapl | SVM | Su(var) | Verni et al., 2000 |
Localization patterns of known heterochromatin components, IP-MS and RNAi screen hits. Top candidates from the localization screen and proteins with a previously known connection to HP1a were imaged at higher resolution and grouped into four categories of heterochromatin localization, based on live imaging in the presence of fluorescently tagged HP1a: broad, narrow, focal, or at the heterochromatin boundary. Localization outside of heterochromatin is also noted. Proteins are sorted by their observed localization patterns. HC = heterochromatin, NR = nucleolar, EC = euchromatin, CP = cytoplasmic.
| Heterochromatic Localization | Other Localization Notes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene Name | Isoform | Reason Investigated | Broad | Narrow | Focal | At HC Boundary | Pan Nuclear | Other | Previous Published Localization | Effect on Variegation |
| Heterochromatin protein 4 | HP4-RA | HP1a IP-MS | X | Kc chromocenter (Greil et al., 2007) | Su(var) (Greil et al., 2007) | |||||
| Heterochromatin protein 5 | HP5-RA¶ | HP1a IP-MS | X | Kc chromocenter (Greil et al., 2007) | Su(var) (Greil et al., 2007) | |||||
| Lysine (K)-specific demethylase 4A | Kdm4A-RA¶ | HP1a IP-MS | X | Kc, S2 and BG3 chromocenter (Colmenares et al., unpublished) | Su(var) (Colmenares et al., unpublished) | |||||
| Suppressor of variegation 3-9 | Su(var)3-9-RA‡,¶ | HP1a IP-MS | X | polytene chromocenter (Schotta et al., 2002) | Su(var) (Reuter et al., 1986) | |||||
| Suppressor of variegation 3-7 | Su(var)3-7-RB†,¶ | literature | X | polytene chromocenter, HC in embryos (Cleard et al., 1997) | Su(var) (Reuter et al., 1990) | |||||
| Lethal hybrid rescue | Lhr-RA/HP3-RA¶ | HP1a IP-MS | X | X | centromeric (Thomae et al., 2013); polytene chromocenter (Brideau et al., 2006); Kc chromocenter (Greil et al., 2007) | Su(var) (this study) | ||||
| Heterochromatin protein 6 | HP6-RA¶ | literature | X | Slight narrow HC enrichment | Kc chromocenter (Greil et al., 2007); polytene chromocenter (Joppich et al., 2009); Kc cells - centromeric (Ross et al., 2013) | Not a mod(var) (Greil et al., 2007); deficiency spanning gene is a Su(var) (Doheny et al., 2008) | ||||
| Oddjob (CG7357) | Odj-RA¶ | HP1a IP-MS | X | X | X | - | Su(var) (this study) | |||
| Su(var)2-HP2 | Su(var)2-HP2-RB | HP1a IP-MS | X | X | X | polytene chromocenter (Shaffer et al., 2002) | Su(var) (Shaffer et al., 2002) | |||
| blanks | blanks-RA* | RNAi screen | X | X | X | Foci outside HC | pan-nuclear (structured) (Gerbasi et al., 2011) | Su(var) (this study); OE mod(var) (Schneiderman et al., 2010) | ||
| CG2129 | CG2129-RA* | RNAi screen | X | X | Foci outside HC | - | RNAi lines were lethal | |||
| FK506-binding protein 1 | FK506-bp1-RA | HP1a IP-MS | X | Foci outside HC | nucleolar based on DAPI-staining (Edlich-Muth et al., 2015) | Non-mod(var) (this study) | ||||
| XNP | XNP-RA¶ | literature | X | X | active genes and satellite DNA near HC in polytenes and imaginal discs (Schneiderman et al., 2009); Broad HC in polytenes (Bassett et al., 2008); Beta-heterochromatin of the X chromosome in polytenes (Emelyanov et al., 2010) | OE mod(var) (Schneiderman et al., 2009); Su(var) (Bassett et al., 2008), (Emelyanov et al., 2010) | ||||
| Suppressor of Under-Replication | SuUR-RA¶ | literature | X | X | polytene chromocenter (Makunin et al., 2002) | mutation is Su(var), extra copy is E(var): (Belyaeva et al., 2003) | ||||
| Hormone receptor 83 | Hr83-RA*,§ | RNAi screen | X | X | NR | - | - | |||
| D1 chromosomal protein | D1-RA¶ | literature | X | Slight narrow HC enrichment | HC (SATI and SATIII) in embryos (Aulner et al., 2002) | Su(var) (Aulner et al., 2002) | ||||
| lethal (3) neo38 | l(3)neo38-RB | RNAi screen | X | FOCI | - | Non-mod(var) (this study); OE mod(var) (Schneiderman et al., 2010) | ||||
| crooked legs | crol-RD | RNAi screen | X | FOCI | nuclear (Mitchell et al., 2008) | Su(var) (this study); OE mod(var) (Schneiderman et al., 2010) | ||||
| ADD domain-containing protein 1 | ADD1-RB | HP1a IP-MS | X | X | Weak broad HC enrichment | polytene chromocenter (Alekseyenko et al., 2014) | Su(var) (Alekseyenko et al., 2014) | |||
| proliferation disrupter | prod-RA¶ | literature | X | X | AATAACATAG in 3rd instar larvae brains (Platero et al., 1998) | - | ||||
| Heterogeneous nuclear ribonucleoprotein at 87F | Hrb87F-RA§ | RNAi screen | X | polytene chromocenter (Piacentini et al., 2009) | Su(var) (Piacentini et al., 2009) | |||||
| Tousled-like kinase | Tlk-RF | RNAi screen | X | 1-2 foci per nuc. Often 1 focus is abutting HP1a | nuclear, but not chromatin bound (Carrera et al., 2003) | Su(var) (this study) | ||||
| RNA and export factor binding protein 1 | Ref1-RA# | HP1a IP-MS | X | Slight HC enrichment | nuclear membrane and nucleoplasm (Buszczak and Spradling, 2006) | - | ||||
| sans fille | snf-RA | RNAi screen | Except nucleolus | nuclear (Flickinger and Salz, 1994) | Su(var) (this study) | |||||
| Hepatocyte nuclear factor 4 | Hnf4-RA | RNAi screen | Except nucleolus | nuclear (Palanker et al., 2009; Gutzwiller et al., 2010) | Su(var) (this study) | |||||
| bicoid-interacting protein 3 | bin3-RA | RNAi screen | X | - | Su(var) (this study) | |||||
| Cullin 4 | Cul4-RA¶ | literature | X | - | - | |||||
| female lethal d | fl(2)d-RA | HP1a IP-MS | X | non-uniform in nucleus (Penn et al., 2008) | Su(var) (this study) | |||||
| jumeau | jumu-RA§ | RNAi screen | X | polytene chromocenter (Strödicke et al., 2000) | Su(var) (Strödicke et al., 2000) | |||||
| La autoantigen-like | La-RA# | HP1a IP-MS | EC | nuclear (Yoo and Wolin, 1994) | Non-mod(var) (this study) | |||||
| Structure specific recognition protein | Ssrp-RA | RNAi screen | NR | nucleolar (Hsu, et al., 1993) | - | |||||
-
*Protein localization is dependent on which terminus of the gene is GFP-tagged and/or cell-type
-
†Stable tagged Kc cell line
-
‡Transient transfection of BG3 cells
-
§Less than 1% of cells expressed the construct
-
#Proteins were only found enriched in one HP1a IP-MS
-
¶Proteins were not tested for colocalization with HP1a in the low-resolution colocalization screen
Additional files
-
Supplementary file 1
GO enrichment analysis of HPips.
135 HPips were analyzed using DAVID for enrichment of GO terms. The GO enrichment category is shown along with the p-value and the HPips that belong to that GO enrichment category. Some redundant GO categories were removed for ease of viewing by selecting a representative GO category for each Functional Annotation Cluster reported by DAVID. Blue GO enrichment categories indicate that the categories belonged to the same Functional Annotation Cluster. GO categories representing ribosomal subunits and structural constituents of cytoskeleton were enriched, but are not displayed since they are considered common contaminants.
- https://doi.org/10.7554/eLife.16096.035
-
Supplementary file 2
Description of imaging features extracted from images.
Using custom Matlab scripts, the number of nuclei per well were counted and 32 imaging features were measured per nucleus and averaged per well. These averaged imaging features combined with the number of nuclei yields an 'imaging signature' for each well.
- https://doi.org/10.7554/eLife.16096.036
-
Supplementary file 3
564 genes putatively down-regulate HP1a.
Gene depletions that led to an increase in HP1a mean and max intensity are listed (HP1a negative regulators).
- https://doi.org/10.7554/eLife.16096.037
-
Supplementary file 4
GO enrichment analysis on all RNAi hits.
All hits were analyzed using DAVID for enrichment of GO terms. The GO enrichment category is shown along with the p-value and the gene that belongs to that GO enrichment category. Redundant GO categories were removed by selecting a representative GO category for each Functional Annotation Cluster as reported by DAVID. Red indicates a GO category that was not significantly enriched but was deemed to be of interest.
- https://doi.org/10.7554/eLife.16096.038
-
Supplementary file 5
A summary of the data presented in this manuscript.
Unless otherwise noted the data presented here was generated as part of this study. Not all proteins that colocalized with HP1a as part of the 'low-resolution' imaging screen were tested for effects on PEV or imaged at higher resolution. If these proteins were previously studied for PEV effects or localization, references are provided. See the main text for details.
- https://doi.org/10.7554/eLife.16096.039
-
Supplementary file 6
A list of primers used in this manuscript.
All sequences are in 5' to 3' orientation; genic sequences are shown in upper case; flanking sequences containing restriction sites are shown in lower case.
- https://doi.org/10.7554/eLife.16096.040





