Unique membrane properties and enhanced signal processing in human neocortical neurons
Figures
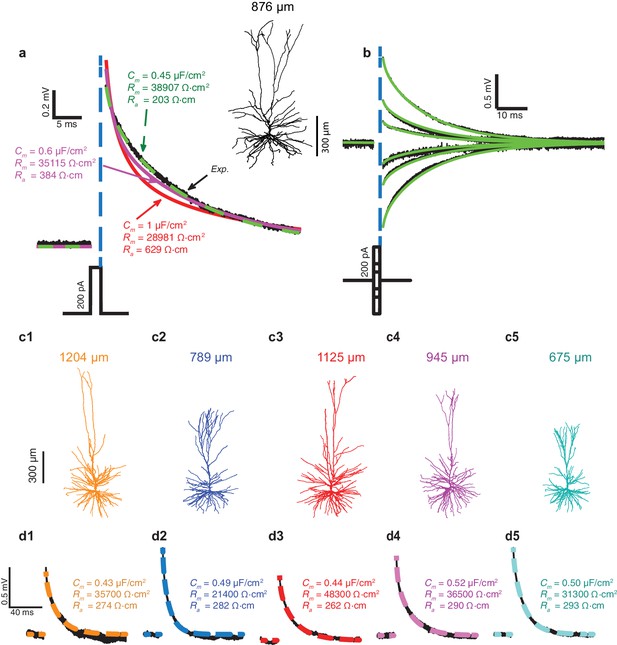
Exceptionally low specific membrane capacitance, Cm, for L2/3 human pyramidal cells predicted using detailed neuron models.
(a) Experimental transient voltage response (upper black trace, Exp.) to a brief somatic depolarizing current pulse (2 ms, 200 pA). The corresponding 3D reconstructed HL2/3 cell from which this voltage transient was obtained is shown in the inset. Color traces depict theoretical transients resulting from injecting the experimental current into the compartmental model of that cell (see Materials and methods). The corresponding cable parameters (Cm, Rm, Ra) are shown for each theoretical transient. Excellent fit was obtained only with Cm value near 0.5 μF/cm2 (green trace), which is half the conventional value of around 1 μF/cm2, obtained for a variety of neuron types, including cortical neurons of other species. (b) Model parameters with low Cm (green traces in a) also match the experimental responses (black) to stimuli of different strength and signs (−200 pA, −100 pA, −50 pA, 50 pA, 100 pA, 200 pA). (c1–c5) Five additional human L2/3 neurons taken from our human neuron database (depth from pia is provided above each cell). (d1–d5) The experimental transient voltage response for the neurons in c1–c5 (black traces), and models fit to the transients (overlapping color traces). In all models, Cm ranged between 0.43 μF/cm2and 0.52 μF/cm2. Patient history for these six cells is described in Table 2.
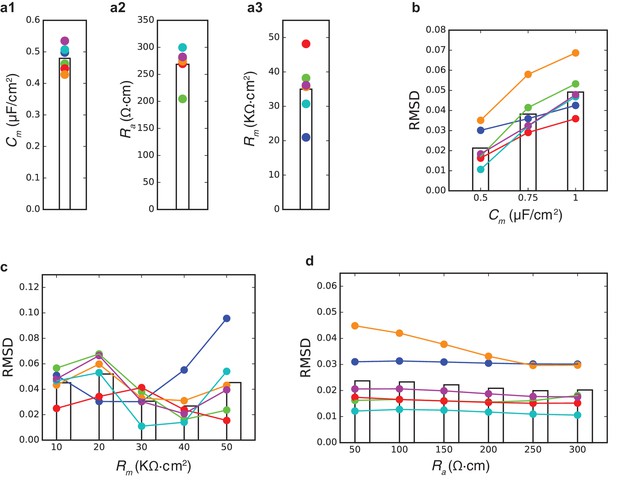
Estimates for passive properties of human L2/3 neurons.
(a1–a3) Model values for the best fits shown in Figure 1 for the three passive parameters Cm, Ra, Rm. Colors match the model colors as in Figure 1. Bar shows the mean value. (b) Root Mean Square Distance of best fits when Cm value was constrained (x-axis) in the optimization procedure. For these simulations, the other passive parameters (Ra, Rm) were free parameters. Note that the smallest RMSD is obtained with Cm = 0.5 µF/cm2. (c) and (d) Similar to b, but when Rm or Ra were constrained. Note the shallow dependence of the RMSD on Ra.
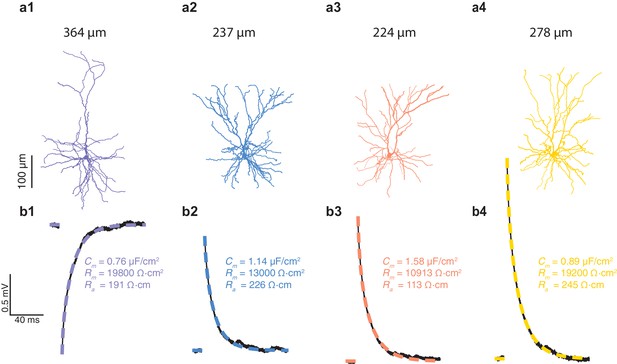
Cable properties for mouse L2/3 pyramidal cells from the temporal cortex.
(a1–a4) Four L2/3 neurons from three mice reconstructed in 3D. (b1–b4) black traces, experimental voltage traces recorded in the four corresponding neurons shown in a1–a4. color traces, best fitted theoretical transients resulting from injecting the experimental current into the compartmental model of that cell. The corresponding cable parameters (Rm, Cm, Ra) are shown on the right.
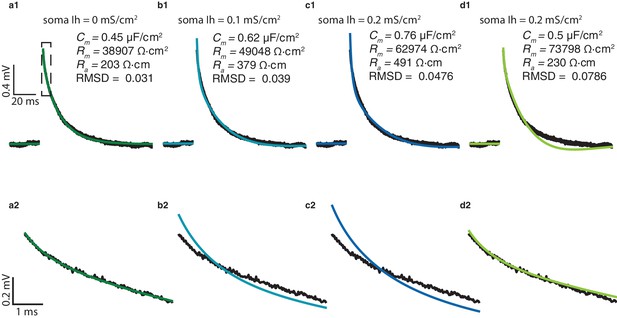
Estimating passive parameters for human L2/3 pyramidal cells when incorporating Ih channels into the model.
(a1) The best fit between experimental transient (black) and theoretical transient (green) for the passive model (no Ih) for the cell shown in Figure 1a. The corresponding cable parameters are shown in the inset. (a2) Zoom in into the first few ms of the transients shown in a1. (b1), (b2), and (c1), (c2); similar to a1, a2 but with somatic Ih density of 0.1 mS/cm2 and 0.2 mS/cm2 respectively, and increased Ih density in the apical tree as in Eq. (4). The theoretical transient is depicted in blue. Note the increase in RMSD and the discrepancy between the experimental and the theoretical transients at the first few ms in both b2 and c2. (d1), (d2) similar to c1, c2 but when Cm was constrained at 0.5 μF/cm2. Note the large discrepancy in the tail of the transient d1 but the excellent match between the model and the experiments in its first few ms.
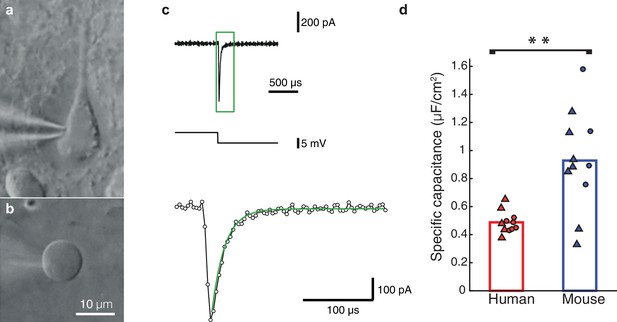
Nucleated-patch measurement of the specific membrane capacitance in HL2/3 neurons directly validated model prediction for the low Cm value in these cells.
(a) Differential Interference Contrast (DIC) image of a human L2/3 pyramidal neuron in an acute brain slice of human temporal cortex targeted for whole-cell patch clamp recording. (b) Nucleated soma patch pulled from the neuron in a. (c) Top trace: Current response of the nucleated patch shown in a to a −5 mV hyperpolarizing step (middle trace). Bottom trace: magnification of the green box in the top trace, showing mono-exponential fit (green line) to the capacitive current from which the total membrane capacitance was extracted. This capacitance was then divided by the surface area of the patch, providing the specific membrane capacitance (see Materials and methods). (d) Summary plot of the specific membrane capacitance from human and mouse. Triangles, Cm values from the nucleated patches. Circles, Cm values obtained from fitting model to experimental transients (as in Figure 1 and Figure 1—figure supplement 2); human (red, n = 11, see details in Table 2), mouse (blue, n = 11). p-val = 0.0021 using students t-test following the KS test for normality.
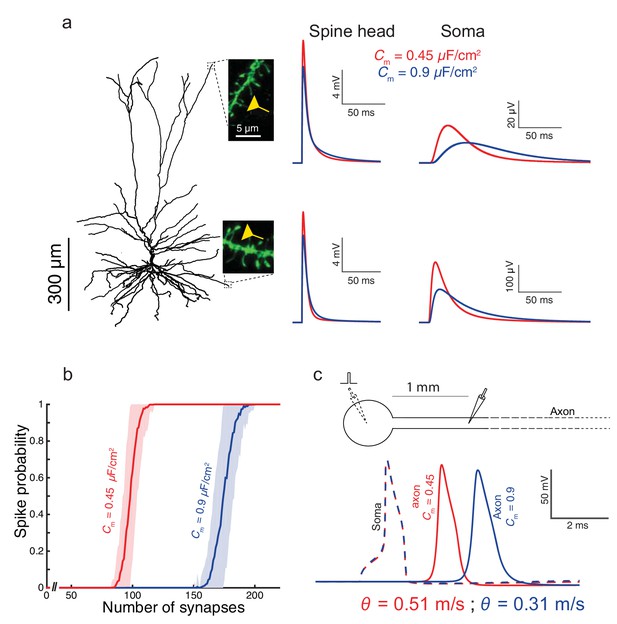
Functional implications of the low Cm value in human L2/3 cortical neurons for signal processing.
(a) The neuron model shown in Figure 1a, receiving an excitatory synaptic input on distal apical dendritic spine (top trace) and on a distal basal dendritic spine (lower traces). The model cell has either Cm = 0.45 μF/cm2 (red traces) or Cm = 0.9 μF/cm2 (blue traces), while the other cable parameters (Ra = 203 Ωcm, Rm = 38,907 Ωcm2) were kept fixed. Excitatory synapses were activated on the head of the modeled dendritic spine (inset, scale bar is 5 μm; see Materials and methods). Note the larger and faster somatic EPSP for the red case. (b) For the cell model shown in a, significantly smaller number of excitatory spinous synapses were required for initiation of a somatic spike when Cm = 0.45 μF/cm2. Synapses were simultaneously activated and distributed randomly over the dendritic tree (see Materials and methods). (c) Top, schematics of soma and axon of the cell modeled in a., the axon had a diameter of 1 µm. Bottom, the velocity (θ) of the axonal spike, measured at 1 mm from the soma, is significantly increased (by about 65%) with Cm = 0.45 μF/cm2 (red spike); the amplitude of the propagated axonal spike is also slightly increased in this case.
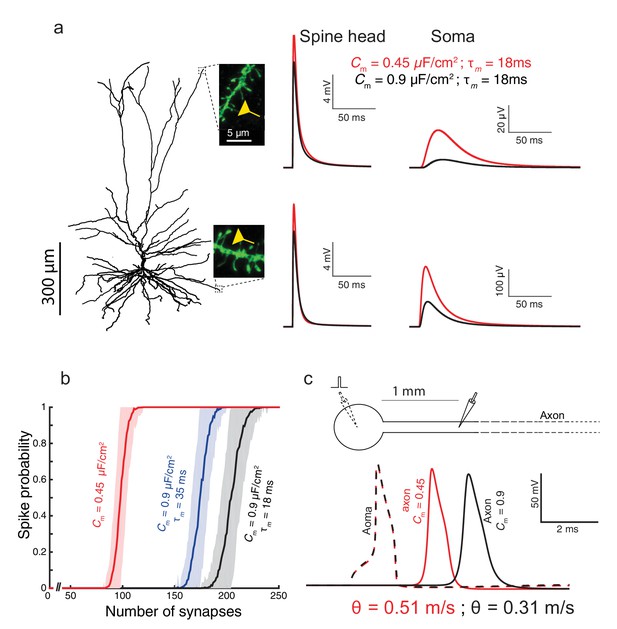
The functional implications of the low Cm value for the case where the membrane time constant, τm, is kept fixed by a corresponding change in Rm for both Cm = 0.45 μF/cm2 and Cm = 0.9 μF/cm2.
(a) As in Figure 3a, but here when Cm = 0.9 μF/cm2 and Rm is halved (Rm = 19,453 Ωcm2). This resulted with smaller differences in the dendritic delay as compared to the case shown in Figure 3a, but with more strongly attenuated EPSPs. (b) As in Figure 3b, but here, due to the smaller EPSPs when Cm = 0.9 μF/cm2, more excitatory inputs are required in order to initiate a somatic spike. (c) Conduction velocity is determined mostly by the axon’s diameter, the axial resistivity, Ra, and the membrane capacitance, Cm (Jack et al., 1975). Consequently, halving Rm had very small effect on spike propagation velocity (less than 1% change between the black traces shown here compared to the blue traces in Figure 3c).
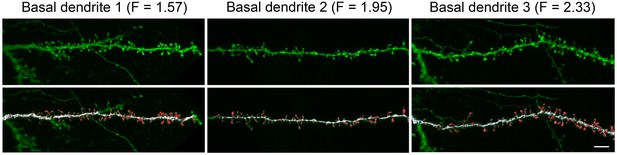
High resolution reconstruction of spines from HL2/3 dendrites.
Top. Three examples of confocal images of basal dendritic branches from post mortem tissue. Bottom. The 3D images were segregated to spines (red) and shafts (white) that were later reconstructed in 3D. The dendritic shaft area and total spines area were calculated from the reconstructions and resulted in a F value for each branch (Equation 2). Scale bar is 5 µm.
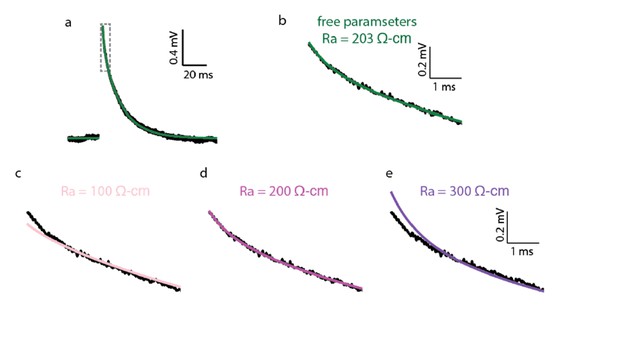
Impact of Ra value on the quality of the fit of the theoretical versus the experimental transients.
(a) Experimental (black trace) and best theoretical fit (green trace) as in Figure 1A. (b) Zoom in on the green square in a. (c–e) Same as in b, but with Ra constrained to 100, 200 and 300 Ω•cm correspondingly. Note the deviation of the fit from the experimental trace in the first couple of ms in c and e. Parameters resulted for the various optimization:
c. Ra = 100 Ω•cm (constrained), Cm = 0.42 μF/cm2, Rm = 41,315 Ω•cm2; d. Ra = 200 Ω•cm (constrained), Cm = 0.46 μF/cm2, Rm = 38,299 Ω•cm2 ;e. Ra = 300 Ω•cm (constrained), Cm = 0.50 μF/cm2, Rm = 36,372 Ω•cm2.
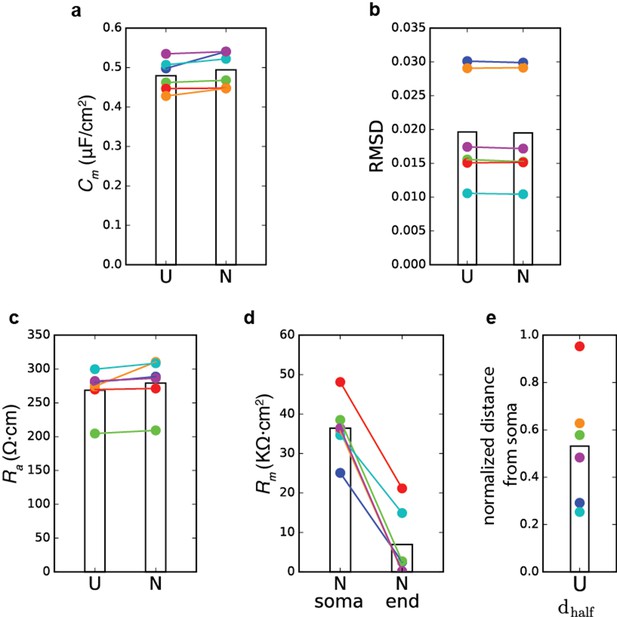
Summary of model parameters for the case of non-uniform Rm.
Model transients were fitted to the experimental voltage transients as in Figure 1. Colors match the colors in Figure 1. (a) Comparison of Cmvalue for the six human models for the case of uniform Rm (U) and the case of non-uniform Rm (N). Bars show the mean value. (b) Root mean square distance between the model responses and the experimental transients. (c) Ra values for the two cases. (d) Rm(soma) versus Rm(end) for the six models, when Rm was assumed to be non-uniform. (e) Normalized for the six models; the maximal dendritic distance in the cells was 1050 ± 178 µm.
Tables
Estimated statistical and systematic errors in the six human L2/3 PCs models shown in Figure 1 as in (Roth and Häusser, 2001). The mean relative statistical error was 6.5% in Cm (range, 4.2-9.2%, n = 5), 8.6% in Rm (range, 4.2-18.8%, n = 5), and 12.8% in Ra (range, 5.3-20.2%, n = 5). In cell #060311, larger errors were found due to several noisy traces. These range of errors in the estimated values of Cm, Rm and Ra, are within the range of values found for the six modeled cells in Figure 1. We also analyzed the case of systematic errors; the errors introduced are shown in Table 3. This leads to, 40% mean relative error in Cm (range, 36.7–58.2%, n = 6), 30.1% in Rm (range, 28.6–34.1%, n = 6), and 53.0% in Ra (range, 47.3–65.4%, n = 6). Thus, even large systematic errors in morphological parameters cannot explain the low Cm (~0.5 µF/cm2) in human L2/3 pyramidal neurons as compared to the typical value ~1 µF/cm2 found in other neurons.
Cell 060303 (Figure 1c2, blue) | Cell 060308 (Figure 1a, green) | Cell 060311 (Figure 1c3, red) | |||||||
|---|---|---|---|---|---|---|---|---|---|
Best fit | S.D. stat | S.D. sys | Best fit | S.D. stat | S.D. sys | Best fit | S.D. stat | S.D. sys | |
Cm (μF/cm2) | 0.488 | 0.045 | 0.183 | 0.452 | 0.026 | 0.196 | 0.441 | 0.248 | 0.257 |
Rm (kΩ∙cm2) | 21.41 | 1.21 | 6.57 | 38.91 | 1.65 | 13.29 | 48.73 | 22.90 | 14.96 |
Ra (Ω∙cm) | 281.78 | 34.12 | 152.05 | 203.23 | 29.31 | 96.18 | 261.97 | 179.3 | 127.22 |
Cell 130303 (Figure 1c1, orange) | Cell 130305 (Figure 1c5, cyan) | Cell 130306 (Figure 1c4, magenta) | |||||||
|---|---|---|---|---|---|---|---|---|---|
Best fit | S.D. stat | S.D. sys | Best fit | S.D. stat | S.D. sys | Best fit | S.D. stat | S.D. sys | |
Cm (μF/cm2) | 0.430 | 0.034 | 0.158 | 0.497 | 0.027 | 0.207 | 0.520 | 0.0219 | 0.236 |
Rm (kΩ∙cm2) | 35.67 | 7.40 | 11.62 | 31.31 | 1.99 | 8.96 | 36.52 | 2.89 | 11.51 |
Ra (Ω∙cm) | 274.44 | 32.24 | 171.63 | 292.95 | 59.04 | 164.46 | 290.28 | 15.41 | 137.46 |
Various parameters for the five patients from which the eleven L2/3 pyramidal cells that were used in this study where taken. CLB, clobazam; CBZ, carbamazepine; FRI, frisium; LCS, lacosamide; LEV, levetiracetam; LTG, lamotrigine; MDZ, midazolam; MTS, mesiotemporal sclerosis; OXC, oxcarbazepine; ZGR, zonegran.
Gender | Age (years) | Age at epilepsy onset (years) | Diagnosis | Seizure frequency (per month) | Antiepileptic drugs (pre-surgery) | Data used in figure |
|---|---|---|---|---|---|---|
Male | 25 | 24 | Tumor | 8 | LEV, CBZ, LCS | Figure 1: a, c2, c3 |
Female | 45 | 23 | MTS (meningitis) | 3 | CBZ, CLB, LTG | Figure 1: c1, c4, c5 |
Female | 40 | 16 | MTS | 7 | ZGR, LTG, FRI, MDZ | |
Male | 43 | 6 | MTS | 7 | LEV, OXC | |
Male | 51 | 4 | unknown etiology | 60 | CBZ, FRI |
Sources of systematic errors.
Error variable | Estimated S.D. |
|---|---|
Scale factor for lengths in the morphological reconstruction | 0.05 |
Additive error in reconstructed diameters (µm) | 0.3 |
Multiplicative error in reconstructed diameters | 0.1 |
Error in spine scale factor, F | 0.4 |
Error in the start point of high density spines (μm) | 30 |
Synaptic properties for Figure 3.
AMPA | NMDA | |
|---|---|---|
0.3 | 3 | |
1.8 | 70 | |
(nS) | 0.7 | 1.4 |
| - | 0.08 |
| - | 1/3.57 |
| - | 1 mM |
Additional files
-
Source code 1
NEURON code for the human models in this work.
- https://doi.org/10.7554/eLife.16553.014





