The novel SH3 domain protein Dlish/CG10933 mediates fat signaling in Drosophila by binding and regulating Dachs
Figures
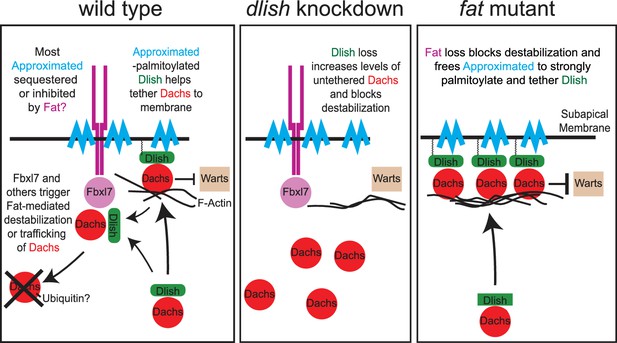
Models of Fat-mediated regulation of subapical Dachs by Fat-inhibited subapical tethering and Fat-stimulated destabilization.
In wild type, Dlish binds Dachs, and when palmitoylated by Approximated helps tether Dachs to the subapical membrane where Dachs can inhibit Warts; Dachs contributes to tethering by binding the F-actin cytoskeleton. Fat binds Approximated and reduces its activity, reducing Dachs tethering. Fat also binds Fbxl7 and other ubiquitin ligases, destabilizing Dachs and trafficking it from the membrane. Dlish promotes the destabilization of Dachs, either indirectly by tethering it near Fat-bound ubiquitin ligases, or directly by helping Dachs complex with those ligases. In dlish mutants both the tethering and destabilization of Dachs are reduced; excess Dachs accumulates in the cytoplasm but no longer inhibits Warts. In fat mutants Approximated-mediated tethering increases and Fat-mediated destabilization of Dachs is lost, increasing subapical Dachs and the inhibition of Warts.
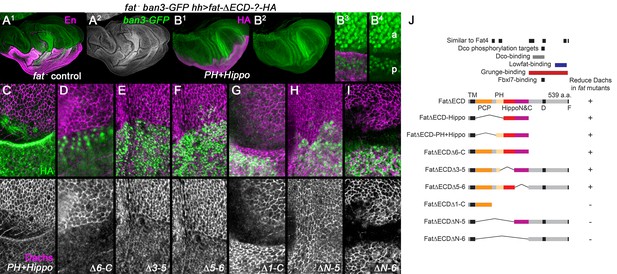
Regulation of Dachs levels by domains in the Fat ICD.
(A1,A2) Control fat; hh-gal4 ban3-GFP wing imaginal disc showing posterior with anti-Engrailed (En). Anterior and posterior are equally overgrown and the Yorkie activity reporter ban3-GFP is expressed at equally high levels in both regions. (B1–I) fat mutant wing imaginal discs expressing FatΔECD-HA constructs in the posterior with hh-gal4, identified using anti-HA. (B1–B4) Reduced overgrowth and ban3-GFP expression by posterior expression of FatΔECD-PH+Hippo. (C–I) Details of anterior-posterior boundary regions showing the posterior response of subapical Dachs to constructs (HA). (J) Summary of constructs and results.
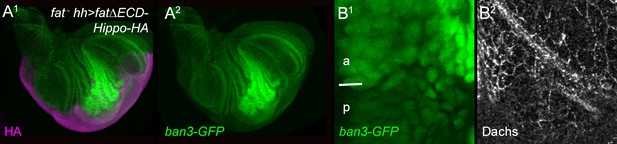
Effect of FatΔECD-Hippo on growth, ban3-GFP and Dachs.
Posterior, hh-gal4-driven expression of FatΔECD-Hippo-HA reduces overgrowth (HA, purple) (A1), ban3-GFP expression (green) (A2,B1), and subapical Dachs (B2).
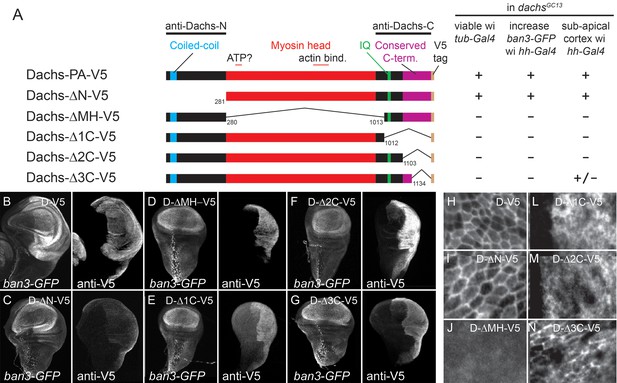
Structure-function analysis of Dachs.
(A) Summary of Dachs-V5 deletion constructs and results of functional assays in dachs mutants. (B–G) Response of the Yki activity reporter ban3-GFP to the expression of Dachs-V5 deletion constructs in dachs mutant background; ban3-GFP is only increased by Dachs-V5 (B) and Dachs-ΔN-V5 (C). (H–N) Localization of Dachs-V5 constructs at subapical focal plane.

Clustal omega alignment of N-termini and C-termini of Drosophila melanogaster Dachs-PA to predicted Dachs homologs.
Shown are domains, the positions of the C terminal deletion breakpoints of the Dachs constructs of Figure 2, and the putative SH3 binding domains of Figure 4D. Tribolium castaneum (Insect) XP_969433.1; Metaseiulus occidentalis (Chelicerate) XP_003739021.1; Limulus polyphemus (Chelicerate) XP_013784597.1; Capitella teleta (Annelid) ELU07103.1; Lottia gigantea (Mollusc) XP_009054577.1; Saccoglossus kowalevskii (Hemichordate) XP_006820318.1; Branchiostoma floridae (Cephalochordate) jgi|Brafl1|227412|e_gw.245.166.1; Nematostella vectensis (Cnidarian) XP_001634718.1; Amphimedon queenslandica (Poriferan) XP_003384558.1; Trichoplax adhaerens (Placozoan) XP_002110197.1
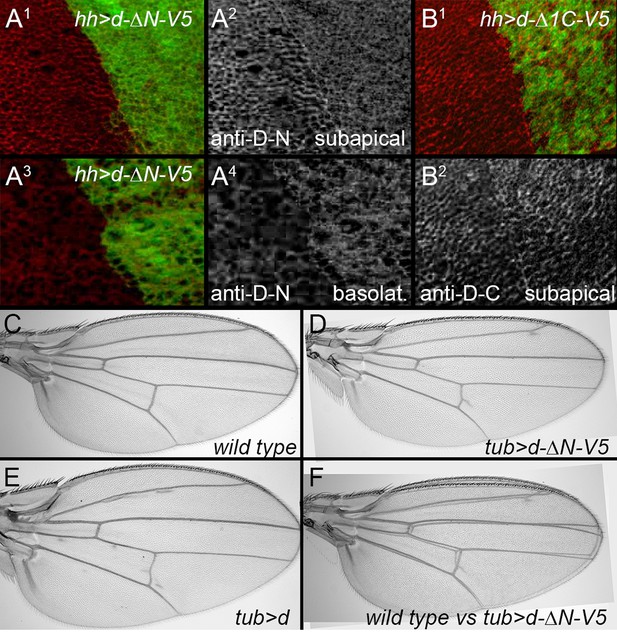
Effects of expressing truncated Dachs in wild type.
(A1–A4) Posterior, hh-gal4-driven expression of UAS-dachs-ΔN (green) slightly reduces the subapical levels (A2), and increases the basolateral, cytoplasmic levels (A4) of endogenous Dachs (recognized by an N-terminal specific anti-Dachs). (B1,B2) Posterior, hh-gal4-driven expression of Dachs-Δ1C (green) does not reduce subapical levels of endogenous Dachs (recognized with a C-terminal specific anti-Dachs). (C–F). tub-gal4-driven expression of UAS-dachs-ΔN slightly reduces wing size (D, F), unlike the overgrowth caused by expression of UAS-dachs (E).
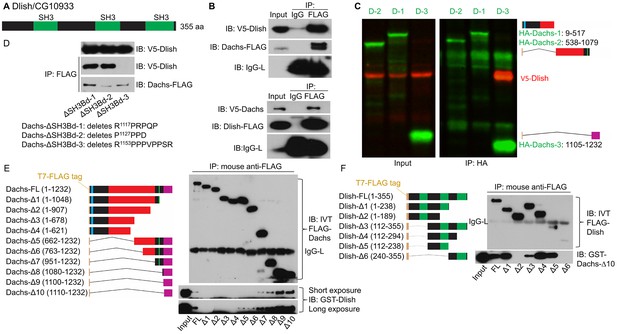
Binding between CG10933/Dlish and Dachs.
(A) Schematic of Dlish domain structure. (B). Reciprocal co-IP between Dachs and Dlish from S2 cells. (C) Co-IP of V5-Dlish from CL8 cells only with the C-terminus of HA-Dachs. (D) Failure of Dlish to co-IP with Dachs from which the third of three candidate SH3 binding domains (SH3Bd) has been removed; the correspondence of SH3Bd-3 with the consensus for type I and type II binding domains in shown in Figure 3—figure supplement 1. (E) Co-IP of GST-purified Dlish only with in vitro-translated (IVT) FLAG-Dachs constructs containing the Dachs C-terminus. (F) Co-IP of GST-purified Dachs C-terminus (Dachs-Δ10) only with those in vitro translated FLAG-Dlish constructs containing the second of its three SH3 domains. See Figure 4—figure supplement 2B for equivalent GST pulldown.
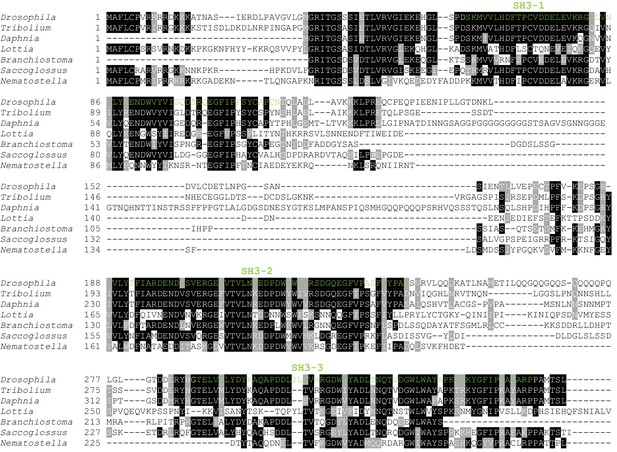
Clustal omega alignment of Drosophila melanogaster Dlish/CG10933 to predicted homologs.
SH3 domains are shown in green. Tribolium castaneum (Insect) XP_970913.2; Daphnia pulex (Crustancean) EFX85167.1; Lottia gigantea (Mollusc) XP_009063502.1;Branchiostoma floridae (Cephalochordate) XP_002590792.1; Saccoglossus kowalevskii (Hemichordate) XP_002732710.1; Nematostella vectensis (Cnidarian) XP_001622159.1.
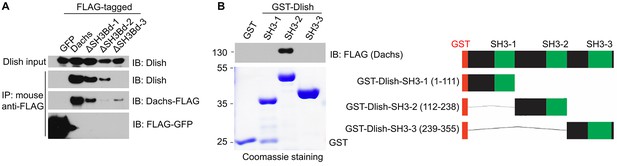
Additional data on binding between Dlish and Dachs.
(A) Failure of Dlish to co-IP with Dachs in which SH3Bd3 has been removed, and with control GFP. (B) GST pulldown of FLAG-tagged Dachs only with the GST-Dlish fragment containing the second of its SH3 domains.
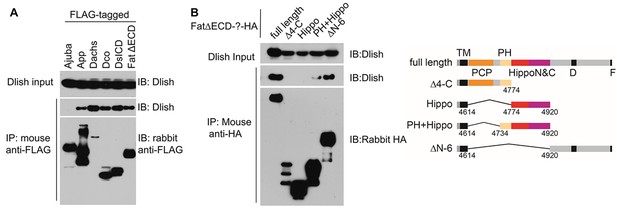
Dlish binding to Fat ICD and Fat pathway members.
(A) Co-IP of Dlish in S2 cells with candidate Fat pathway proteins. Failure to bind Ajuba serves as a negative control. (B) Co-IP of Dlish from S2 cells with FatΔECD, FatΔECD-PH+Hippo and FatΔECDΔN-6, but not FatΔECDΔ4-C or FatΔECD-Hippo.
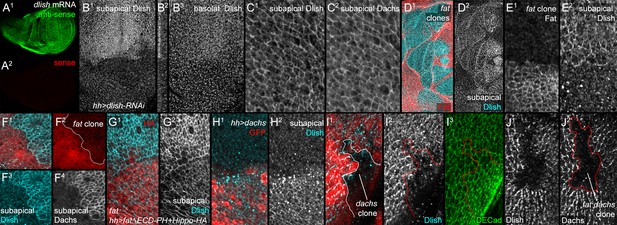
Localization of Dlish and its regulation by Fat and Dachs in wing imaginal discs.
(A1,A2) In situ hybridization to wing imaginal disc with anti-sense (A1) and sense (A2) probes. (B1–B3) hh-gal4 UAS-dlish-RNAi disc stained with anti-Dlish. Dlish remaining in anterior is concentrated subapically (B1, left in cross-section in B2) and diffusely in the basolateral cytoplasm. (C1,C2) Closeup showing overlap of Dlish and Dachs in the subapical cell cortex. (D1–F4) Homozygous fatfd clones, shown by absence of anti-Fat stain (D1,E1) or RFP (F1,F2). Dlish is increased in the subapical cell cortex, coincident with Dachs (F4). (G1,G2) Reduction of subapical Dlish in fat mutant disc by posterior, hh-gal4-driven expression of UAS-fatΔECD-PH+Hippo-HA (anti-HA). (H1,H2) Increase in subapical Dlish by posterior, hh-gal4-driven expression of UAS-dachs (posterior identified with UAS-driven GFP, red) (I1–I3) dachsgc13 homozygous clone (outline) marked by absence of nuclear RFP (I1). Subapical Dlish is decreased, while DE-cadherin (I3) is unchanged. (J1,J2) fatfd dachsgc13 homozygous clone marked by absence of Dachs (outline in J2). Subapical Dlish (J1) is decreased.
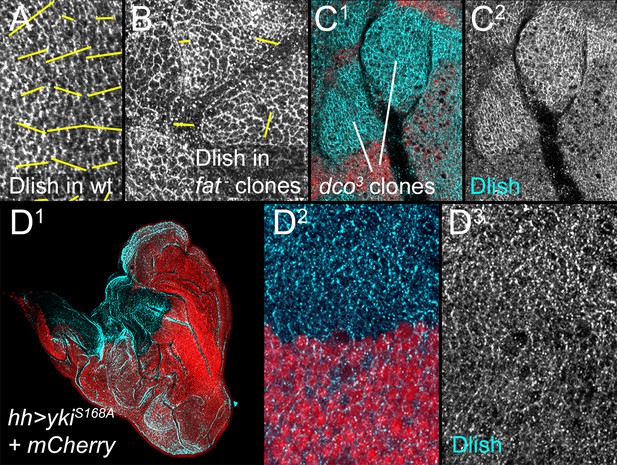
Additional data on the regulation of Dlish localization.
(A,B) Nematics (yellow lines) of Dlish polarity in wild type (wt) disc (A) compared with the lack of consistent nematics in fatfd homozygous clones (B). (C1,C2) Increase in subapical Dlish (blue, white) in homozygous dco3 clones (lack of red RFP). (D1–D3) Strong activation of downstream Yki activity and overgrowth by driving UAS-ykiS168A in posterior with hh-gal4 does not increase subapical Dlish.
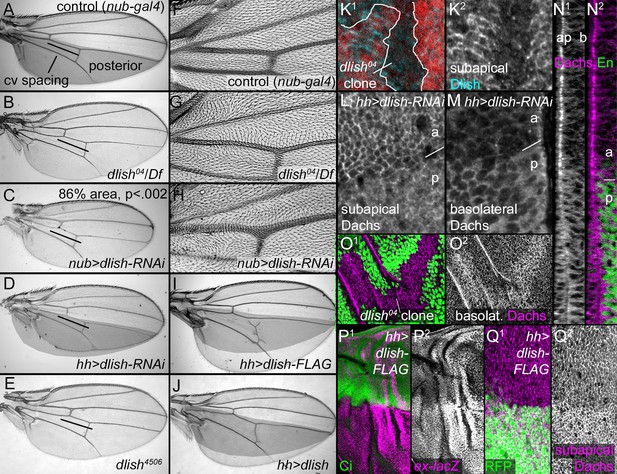
Dlish regulates Dachs function, localization and levels.
(A–J) Adult wings. (A,F) Control, phenotypically wild type nub-gal4 wing. The posterior compartment is marked in grey, and the bar marks the distance between anterior and posterior crossveins. (B,G). dlish04/Df(2R)Exel7150 wing with reduced crossvein spacing (B) and proximal hair PCP defects (G). (C,H) nub-gal4 UAS-dlish-RNAi (VDRC) wing with reduced area (significant by Student’s T and Whitney-Mann tests; see Figure 7—source data 1 for data analysis) and PCP defects (H). (D) Posterior-specific, hh-gal4-driven UAS-dlish-RNAi (VDRC) reduces posterior size compared to grey control area from A. (E) dlish4506 homozygous wing showing reduced crossvein spacing; hair PCP defects are similar to dlish04 (detail not shown). (I,J) Posterior, hh-gal4-driven expression of UAS-dlish-FLAG (I) or UAS-dlish (J) increases posterior size compared to grey control area from A. (K1,K2) Reduction of subapical anti-Dlish staining in dlish04 clone marked by absence of GFP (red). Because the GFP is in nuclei, the clone marker image is from the more basal, nuclear focal plane and slightly out of register with subapical Dlish. (L–N2) Posterior, hh-gal4 driven expression of UAS-dlish-RNAi decreases cortical, subapical Dachs (L) and increases basolateral, cytoplasmic Dachs (M); N1 and N2 show the Dachs changes in the same cross-section Z series projection that shows Dlish in Figure 6B2, with Engrailed (En) marking the posterior (p) versus anterior (a). (O1,O2) dlish04 homozygous clone, marked by absence of GFP, increases basolateral, cytoplasmic Dachs. (P1–Q2) Posterior, hh-gal4-driven expression of UAS-dlish-FLAG increases expression of the Yorkie activity reporter ex-lacZ (P1,P2) and increases subapical Dachs (Q1,Q2); anterior marked with Ci (P1) or posterior with GFP (Q1).
-
Figure 7—source data 1
Data and analysis for comparison of adult wing sizes between nub-gal4 and nub-gal4 UAS-dlish-RNAi for Figure 7C.
- https://doi.org/10.7554/eLife.16624.016
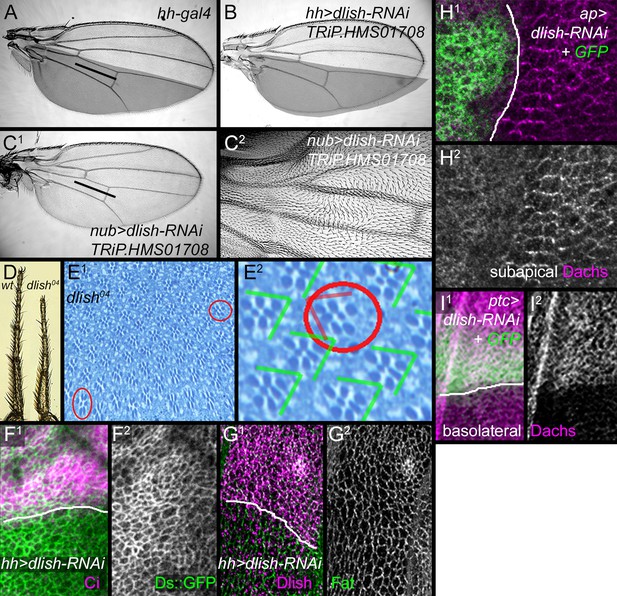
Additional data on dlish phenotypes.
(A) Control hh-gal4 wing. (B–C2) Wing phenotypes from expression of TRiP.HMS01708 UAS-dlish-RNAi. (B) Reduced size of posterior compartment using hh-gal4. (C1,C2) Reduced crossvein spacing and proximal hair PCP defects with nub-gal4. (D) Comparison of tarsal leg segments between wild type and dlish04 homozygote. (E1,E2) Occasional ommatidial PCP reversals (red circles) in dlish04 homozygote retina. (F,G) Unchanged Ds::GFP (F1,F2) or anti-Fat (G1,G2) after posterior, hh-gal4-driven expression of UAS-dlish-RNAi (VDRC); anterior marked with Ci (F1) or Dlish (G1). (H1,H2) Dorsal, ap-gal4-driven UAS-dlish-RNAi (VDRC) reduces subapical cortical Dachs; dorsal is marked with GFP. (I1,I2) Anterior, ptc-gal4-driven UAS-dlish-RNAi (VDRC) increases basolateral cytoplasmic Dachs; anterior is marked with GFP.
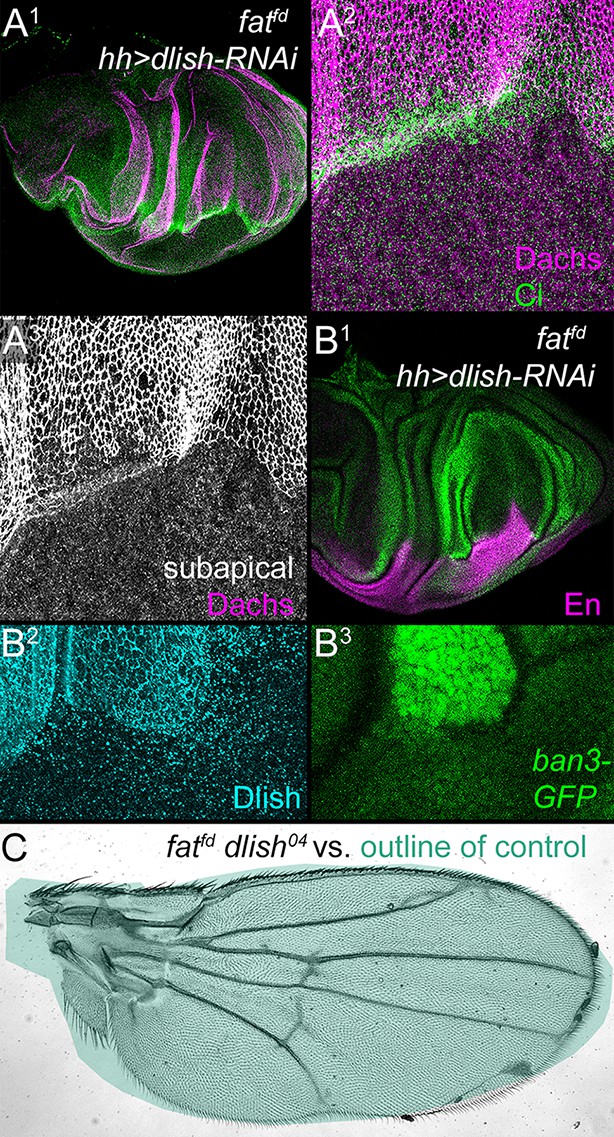
Reduced Dlish function suppresses the overgrowth and Dachs upregulation of fat mutants.
(A1–B3) fatfd discs with posterior, hh-gal4-driven expression of UAS-dlish-RNAi. Anterior marked with Ci (A1,A2), or posterior with En (B1) or Dlish loss (B2). Overgrowth is reduced in the posterior (A1,B1, compare with Figure 2A), as is subapical Dachs (A2,A3) and the Yorkie activity reporter ban3-GFP (B1,B3). (C) fatfd dlish04 double mutant wing is abnormally patterned but nearly-normally sized when compared with the green outline of a control wing from Figure 7A. Unlike the 100% lethality of fatfd, 50–60% of fatfd dlish04 survive to adulthood (44/230 from self-crossed dlish04 fatfd / CyO-TM6,Tb vs. 77/230 expected from self-crossed non-lethal / CyO-TM6,Tb).
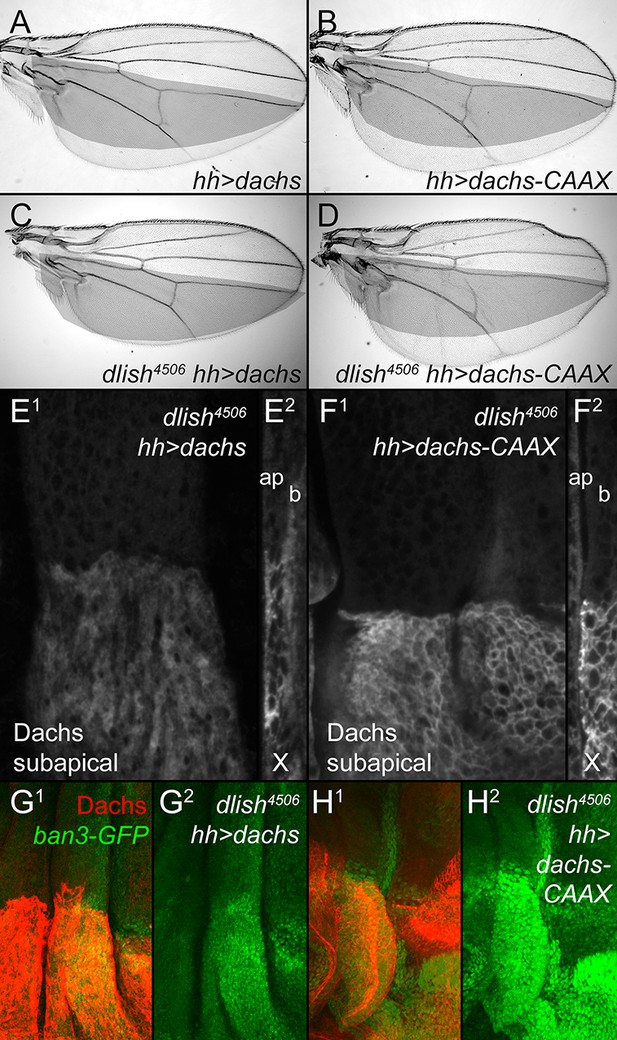
Dlish is required for the function of overexpressed Dachs but not Dachs-CAAX.
(A,B) Similar overgrowth induced by posterior, hh-gal4-driven expression of UAS-dachs-V5 (A) and UAS-dachs-CAAX (B). (C,D) Overexpression in dlish4506 homozygote largely blocks overgrowth induced by UAS-dachs-V5 (C) but not UAS-dachs-CAAX (D). (E,F) Comparison of anti-Dachs staining in dlish4506 wing discs after posterior overexpression of UAS-dachs-V5 (E) or UAS-dachs-CAAX (F), showing subapical focus (E1,F1) or z-series cross-section (E2,F2, apical left). Cortical localization of Dachs-V5 is lost (compare with Figure 3H), but of Dachs-CAAX is retained. Discs were fixed and stained in parallel and imaged with identical settings. (G,H) Comparison showing weaker posterior upregulation of the Hippo activity marker ban3-GFP in dlish4506 wing discs by posterior overexpression of UAS-dachs-V5 (G) than by UAS-dachs-CAAX (H), imaged using the same confocal settings for GFP.
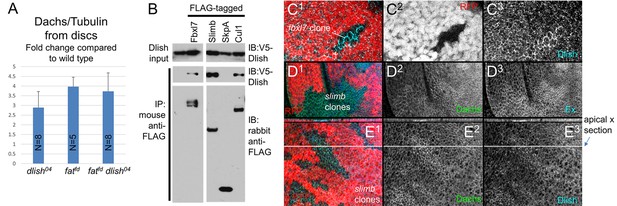
Regulation of Dachs and Dlish levels.
(A) Dachs levels in extracts from wing imaginal discs, normalized to α-Tubulin levels and compared to wild type from the same trial. All Dachs changes were significantly different from wild type, but not from each other. Dachs levels in dlish fat double mutants are significantly lower than the predicted additive effects of dachs and fat (two-tailed p<0.05 using the Wilcoxon Rank Sum test, p=0.000036 using a single sample T test; see Figure 10—source data 1 for data and analysis). (B) Co-IP of Dlish in S2 cells with F-box proteins Fbxl7 and Slimb and with Cullin 1, but not SkpA, from a single exposure of a single blot. See Figure 10—figure supplement 1 for more examples. (C1–E3) Subapical proteins in mutant clones, marked by absence of RFP. RFP is from the nuclear, more basal focal plane, slightly out of register with subapical clone boundaries. (C1–C3) Increased subapical Dlish in fbxl7 clone. (D1–D3) Increased subapical Dachs and Expanded (Ex) in slimb1 clones. (E1–E3) Increased subapical Dachs and Dlish in slimb1 clones; subapical cross sections from marked line are shown above each frame.
-
Figure 10—source data 1
Data and analysis of changes in Dachs in imaginal disc extracts for Figure 10A.
Tests shown are two-tailed. The slight increase in dlish04 fatfd double mutants in comparison to dlish04 did not reach statistical significance, and was significantly lower that the added average increases of dlish04 and fatfd (Wilcoxon Rank Sum test or single sample T test to medians or means of 6.86). The N for dlish04 fatfd is too low to obtain an exact p value using the Wilcoxon test, but still predicts a significance cutoff.
- https://doi.org/10.7554/eLife.16624.021
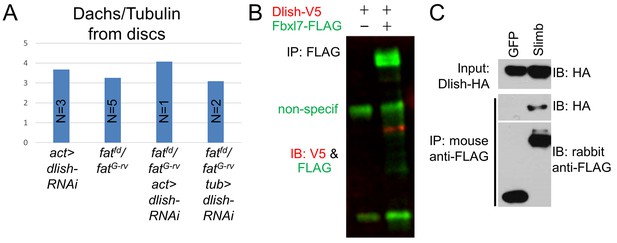
Additional data on Dachs levels and Dlish binding.
(A) Dachs levels from imaginal discs with ubiquitously expressed dlish-RNAi (VDRC) and fatfd/fatG-rv. tub-gal4 drives stronger expression than act-gal4. (B,C) Co-IP of Dlish with Fbxl7 in CL8 cells (B) and Slimb in S2 cells (C).
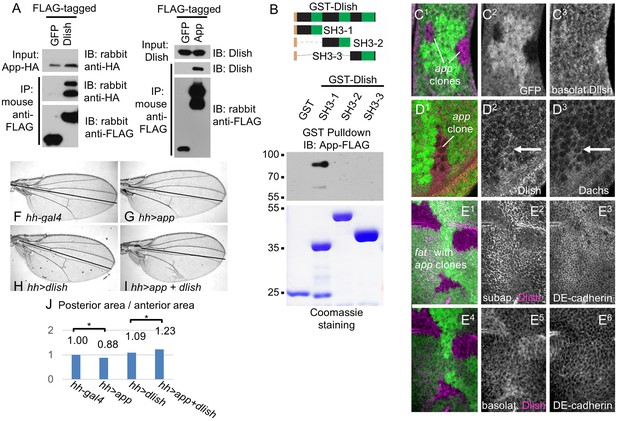
Dlish binds to and is regulated by App.
(A) Reciprocal co-IP of App-HA by Dlish-FLAG and Dlish by App-FLAG. (B) Pulldown of S2 cell-generated App-FLAG by a GST-Dlish construct containing its first SH3 domain. (C1–D3) Increased cytoplasmic Dlish and Dachs in homozygous app clones marked by absence of GFP. (E1–E6) app clones marked by absence of GFP in a fat mutant wing disc. Subapical Dlish levels decrease (E2) and basolateral, cytoplasmic levels increase (E5), while control DE-cadherin is unchanged (E3,E6). (F–J) Posterior, hh-gal4-driven expression of UAS-app decreases the wing area in a wild type wing, but increases the overgrowth induced by UAS-dlish (untagged). Brackets show p<0.01 by a single-tailed Student’s T and Whitney-Mann tests; data and analysis shown in Figure 11—source data 1.
-
Figure 11—source data 1
Data and analysis of wing size change after hh-gal4-driven expression of UAS-app, UAS-dlish (untagged), or both, for Figure 11J.
- https://doi.org/10.7554/eLife.16624.024
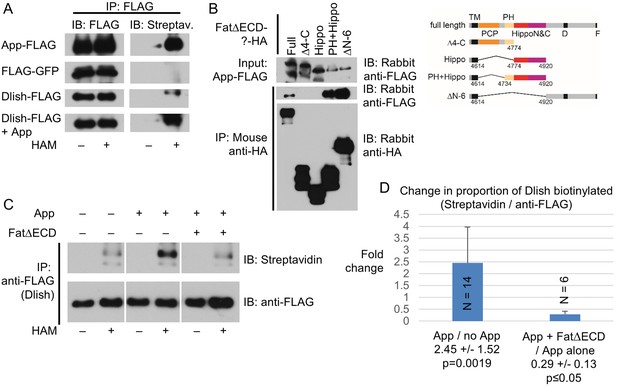
Dlish palmitoylation is regulated by App and the Fat ICD.
(A) ABE assay for Dlish palmitoylation in S2 cells. All anti-FLAG or streptavidin stained lanes are from the same exposure of a single blot. HAM increases streptavidin labeling of Dlish and positive control App, but not negative control GFP. Presence of additional App in Dlish-expressing cells increases labeling. (B) Co-IP of App-FLAG with HA-tagged domains of the Fat ICD co-expressed in S2 cells. (C) ABE assay for Dlish palmitoylation in S2 cells. All anti-FLAG or streptavidin-stained lanes are from the same exposure of a single blot. Presence of additional App increases labeling, but the presence of App and FatΔECD reduces labeling compared with App alone. (D) Quantification of the changes in the proportion of Dlish that is biotinylated, expressed as the average fold-change between two different conditions in individual trials. The presence of additional App significantly increases Dlish labeling compared with no added App; the presence of App and FatΔECD significantly decreases Dlish labeling compared with App alone. p values are from two-tailed Wilcoxon Rank Sum tests to a hypothetical median of 1. For data and Wilcoxon and T test analyses see Figure 12—source data 1.
-
Figure 12—source data 1
Data and analysis of ABE assays for Figure 12D.
Strength of streptavidin binding and anti-FLAG binding was quantified from western blots. To adjust for differences between each trial’s overall staining levels, the experimental streptavidin/anti-FLAG numbers were normalized to the control condition for the same trial, and the number expressed as a fold change. Comparisons were to a hypothetical median of 1 using the two-tailed Wilcoxon Rank Sum test, and to a hypothetical mean of 1 using a two-tailed single-sample T test. The N for App + FatΔECD / App alone is too low to obtain an exact p value using the Wilcoxon test, but still predicts a significance cutoff.
- https://doi.org/10.7554/eLife.16624.026
Tables
Yeast two-hybrid screen with Dachs C-terminus
| 17 hits: |
|---|
| CG10933- SH3 domain protein |
| 2 hits: |
| zld- Zinc finger protein |
| 1 hit each: |
| AOX1- Aldehyde oxidase 1 |
| bc10- Bladder cancer associated protein (BLCAP) homolog |
| bent- Myosin light chain kinase/Titin family (single Ig fragment) |
| CG7220- UBE2w-like E2 Ubiquitin-conjugating enzyme |
| Maf1- RNA polymerase III repressor |
| Strn-Mlck- Myosin light chain kinase/Titin family (single Ig fragment) |
| Tep2- Thioester-containing protein 2 |





