A Cryptochrome 2 mutation yields advanced sleep phase in humans
Figures
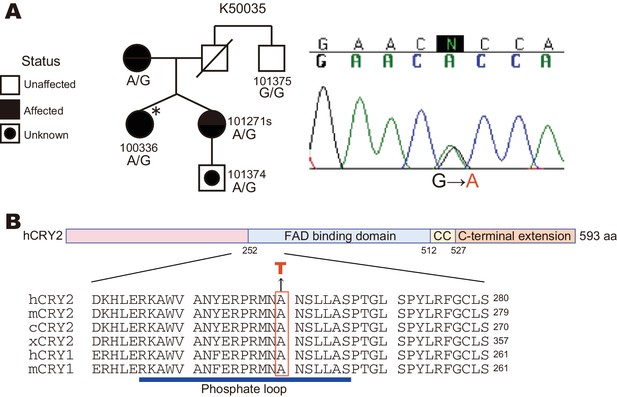
A CRY2 mutation in FASP kindred 50035.
(A) Pedigree of the family (kindred 50035) segregating the CRY2 mutation (A260T). Circles and squares represent women and men, respectively. An asterisk marks the proband. A missense mutation from G to A causes an amino acid conversion from Alanine to Threonine at position 260. (B) Amino acid alignment around the mutation site. The A260T mutation is located in the N-terminal portion of the FAD binding domain in CRY2. This residue is highly conserved among vertebrate species. CC denotes a Coiled-Coil sequence.
-
Figure 1—source data 1
Summary of sleep phenotype of human subjects.
Subject IDs correspondence to numbers in Figure 1A.
- https://doi.org/10.7554/eLife.16695.004
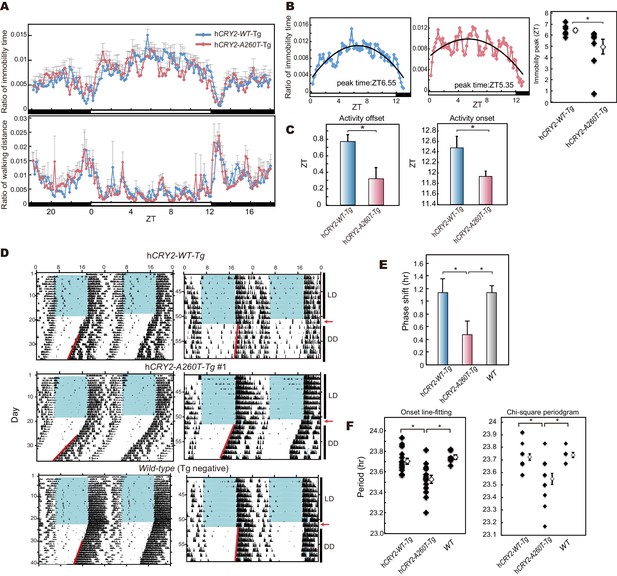
hCRY2-A260T mice have advanced phase of sleep-wake behavior in a light-dark cycle and a shortened circadian period in constant darkness.
(A) Mouse movement was tracked by an infrared video camera in LD. The ratio of immobilization time to total daily immobilization time (upper panel) and the ratio of walking distance to total daily distance (bottom panel) were plotted every 10 min. Data are shown as means with SEM (n = 8 for hCRY2-WT and hCRY2-A260T). (B) Peak time of immobility was measured by fitting a quadratic function to data from ZT0 to 13. Representative examples of curve fitting for hCRY2-WT and hCRY2-A260T are shown here. Data are shown as means with SEM (n = 8 for hCRY2-WT and hCRY2-A260T, *p<0.05 by Student’s t-test). (C) Onset and offset of locomotor activity. Data are shown as means with SEM (n = 8 for hCRY2-WT and hCRY2-A260T). (D) Actograms of wheel-running activity for hCRY2-WT, hCRY2-A260T, and littermate transgene-negative mice. The blue shadows indicate periods when the lights were on. Red lines were fitted to activity onset using ClockLab analysis software. (E) Phase-shifts in response to a 30-min light exposure at ZT14 indicated by red arrows in (D). *p<0.05 by Tukey’s test (n = 7 for hCRY2-WT, n = 11 for hCRY2-A260T, n = 10 for WT). (F) The distribution of period measurements for BAC transgenic mice and transgene negative controls. Period was determined by line fitting of activity onset and chi-square periodogram from day 7 to day 19 in DD. *p<0.05 (n = 15 for hCRY2-WT, n = 14 for hCRY2-A260T and n = 7 for WT)
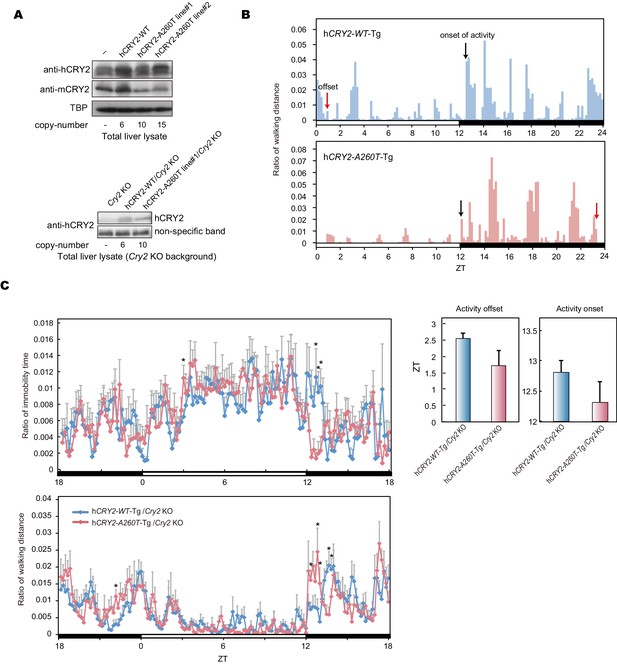
Locomoter activity of h CRY2 BAC Tg mice.
(A) CRY2 expression in total liver lysate at ZT18. CRY2 protein was detected by human CRY2 antibody (Santa Cruz) and mouse CRY2 antibody (Hirano et al., 2013). Copy number was determined by real-time PCR using common sequences in mouse and human Cry2 genes. (B) Representative profiles of locomotor activity measured by video recording. Activity onset (black) and offset (red) are indicated by arrows in the figures. The time of onset and offset for all animals of respective genotypes are averaged and plotted in Figure 2C. (C) Locomotor activity and resting behavior of transgenic mice on a mCry2 knockout background. Mouse movement was tracked by an infrared video camera in LD. The ratio of immobilization time to total daily immobilization time (upper panel) and the ratio of walking distance to total daily distance (bottom panel) were plotted every 10 min. Data are shown as means with SEM (n = 6 for hCRY2-WT/Cry2 KO, n = 6 for hCRY2-A260T/Cry2 KO). The average of activity offset and onset in LD 12:12 are shown (n = 6 for hCRY2-WT/Cry2 KO, n = 6 for hCRY2-A260T/Cry2 KO).
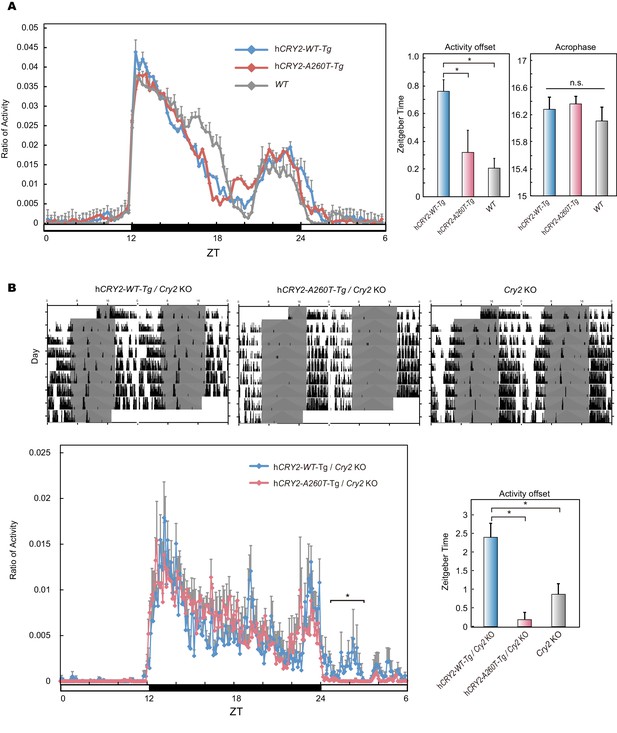
Wheel-running activity of hCRY2 BAC Tg mice.
(A) Activity profiles of transgenic mice in LD 12:12. Ratio of wheel-running counts to total daily counts are plotted every 20 min (n = 18 for hCRY2-WT, n = 14 for hCRY2-A260T). The average of activity offset in LD 12:12 are shown in the right graph. *p<0.05 by Tukey’s test (n = 19 for hCRY2-WT, n = 19 for hCRY2-A260T and n = 7 for WT). (B) Actograms and activity profiles of transgenic mice on a mCry2 knockout background in LD 12:12. Ratio of wheel-running counts to total daily counts are plotted every 5 min (n = 5 for hCRY2-WT, n = 5 for hCRY2-A260T). The average of activity offset in LD 12:12 are shown in the right graph. *p<0.05 by Tukey’s test (n = 5 for hCRY2-WT/mCry2 KO, n = 5 for hCRY2-A260T/mCry2 KO and n = 6 for mCry2 KO).
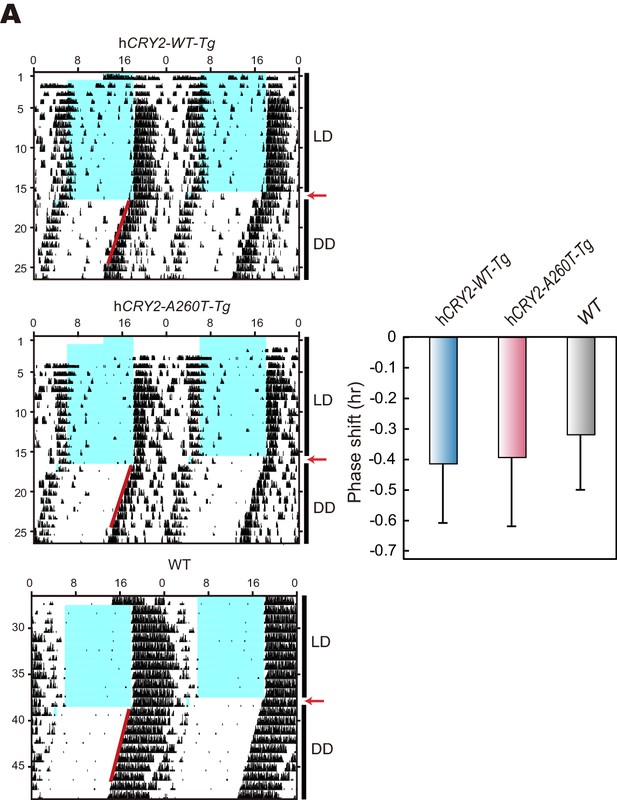
Light phase-shift of hCRY2 BAC Tg mice at ZT22.
(A) Phase-shift in response to 30-min light exposure at ZT22. Mice were exposed to a 30-min light pulse at ZT22 (indicated by red arrows) and released into DD after the light pulse. The blue shadows indicate periods when the lights were on. Red lines were fitted to activity onset using ClockLab analysis software. Phase-shift was determined by line-fitting to activity onset (n = 6 for hCRY2-WT, n = 7 for hCRY2-A260T and WT).
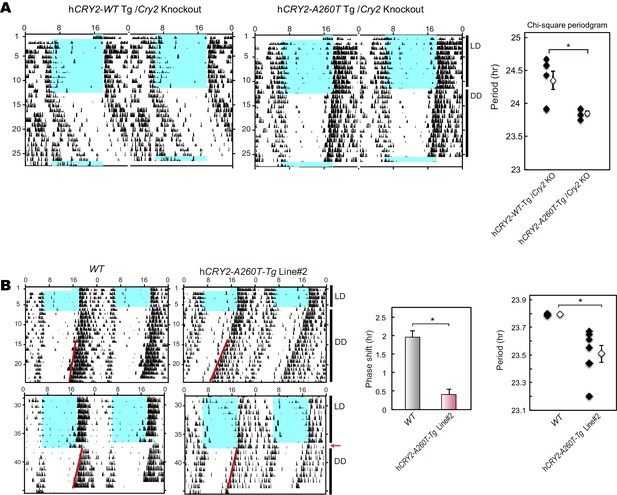
Wheel-running activity of hCRY2Cry2 BAC Tg mice on a m KO background and another BAC Tg mice line.
(A) Circadian period of the transgenic mice on a mCry2 knockout background. Period was determined by chi-square periodgram from day 7 to day 14 in DD. *p<0.05 by Welch’s test (n = 6 for hCRY2-WT/mCry2KO and n = 5 for hCRY2-A260T/mCry2KO). (B) Circadian period and phase-shift of hCRY2-A260T mice line#2. Representative actograms of wheel-running activity of hCRY2-A260T line#2 and littermate controls (WT) are shown. The blue shadows indicate periods when the lights were on. Red lines were fitted to activity onset using ClockLab analysis software. Phase-shifts shown here are by 30-min light exposure at ZT14 (left panel, n = 7 for hCRY2-A260T line#2, n = 3 for WT). Period was determined by line fitting of activity onset from day 7 to day 19 in DD. *p<0.05 by Student’s t-test (right panel, n = 7 for hCRY2-A260T line#2, n = 3 for WT).
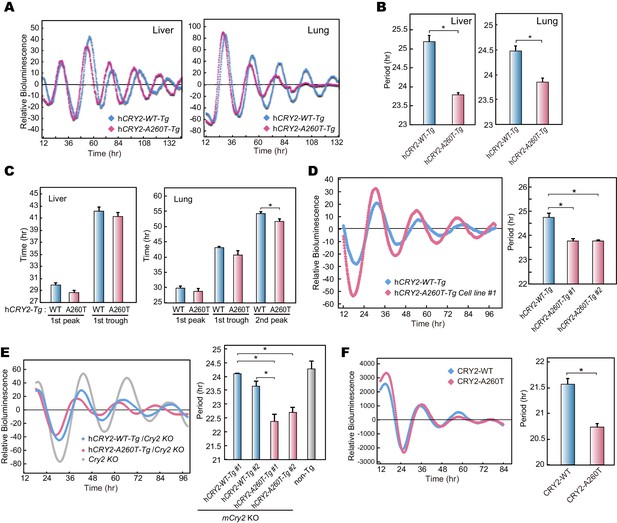
Bioluminescence rhythms in tissue cultures from hCRY2-A260T mice and CRY2-A260T stable cell lines.
(A) Representative rhythms of PER2::LUC bioluminescence in the liver and lung. Data were detrended by subtracting the 24 hr average of bioluminescence. (B) Period measurements of the bioluminescence rhythms in liver and lung tissues. Data are shown as means ± SEM (n = 4 for liver and hCRY2-WT lung, n = 3 for hCRY2-A260T lung, *p<0.05 by Student’s t-test). (C) Peak and trough time of PER2::LUC bioluminescence rhythms of mouse liver and lung tissues. Data are shown as means ± SEM (n = 4 for liver). Data are shown as means ± SD (n = 2 to 4 for lung, *p<0.05 by Student’s t-test). (D) Representative rhythms of PER2::LUC bioluminescence in MEFs from hCRY2 transgenic mice. Data were detrended by subtracting the 24 hr average of bioluminescence. Period lengths of the bioluminescence rhythms are shown as means ± SEM (n = 4, *p<0.05 by Tukey’s test). (E) Representative examples of bioluminescence rhythms of mBmal1-luc in MEFs from transgenic mice on a mCry2 knockout background. Cells were transfected with mBmal1-luc vector 24 hr before the recording. Data were detrended by subtracting the 24 hr average of bioluminescence. Period lengths of the bioluminescence rhythms in the stable cell lines are shown as means ± SEM (n = 4, *p<0.05 by Games-Howell test). (F) Representative examples of bioluminescence rhythms of Bmal1-luc in NIH3T3 cells stably expressing FLAG-CRY2-WT or FLAG-CRY2-A260T. Data were detrended by subtracting the 24 hr average of bioluminescence. Period lengths of the bioluminescence rhythms in the stable cell lines are shown as means ± SEM (n = 3, *p<0.05 by Student’s t-test).
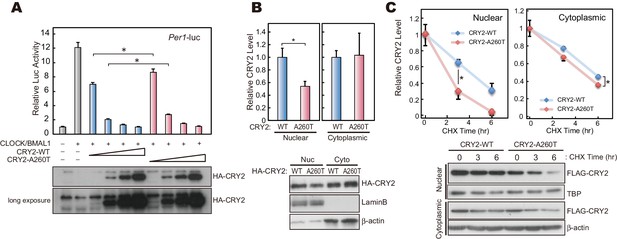
CRY2-A260T is less stable, particularly in nuclei.
(A) Luciferase activity driven by mouse Per1 E-box in HEK293 cells. 2, 5, 10 and 20 ng of hCRY2 expression vector (WT or A260T) was transfected into cells cultured in a 24-well plate. Luciferase activity was normalized to Renilla luciferase activity. Data are shown as means with SEM (n = 4, *p<0.05 by Student’s t-test). (B) Protein levels of CRY2-WT and CRY2-A260T in the nuclear and cytosolic fractions of HEK293 cells. Data are shown as means with SEM (n = 3, *p<0.05 by Student’s t-test). (C) Degradation assay of CRY2 protein in HEK293 cells. Forty-eight hours after transfection, cells were treated with 100 μg/ml CHX and fractionated into the nuclear and cytosolic fractions. CRY2 protein levels at the starting point (t = 0 hr) were normalized to 1. Data are shown as means with SEM (n = 3*, p<0.05 by Student’s t-test).
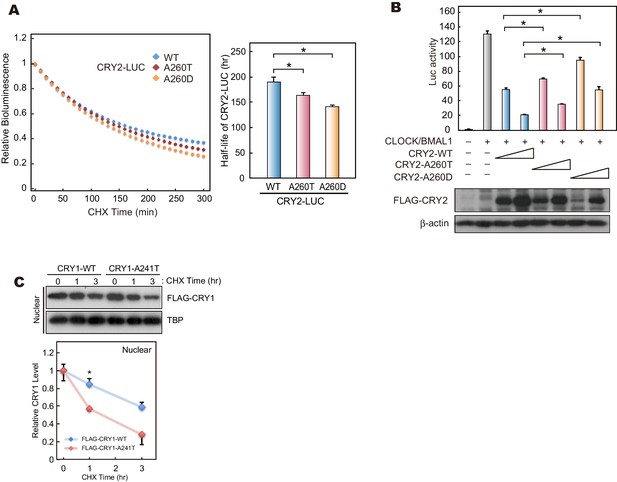
The effects of mutations at A260 on protein stability, repressor activity.
(A) Twenty-four hours after transfection, the culture media was changed to recording media containing 100 µg/ml CHX, and bioluminescence of CRY2-LUC (WT, A260T or A260D) was recorded continuously. Bioluminescence normalized to the start point was fitted to an exponential curve to determine half-life of CRY2-LUC (right panel). Data are shown as means ± SEM (n = 4, *p<0.05 by Tukey’s test). Left panel shows representative bioluminescence decay of CRY2-LUC. (B) Luciferase activity driven by mouse Per1 E-box. 10 or 25 ng of CRY2 construct was transfected to HEK293 cells cultured in 24-well plate. Luciferase activity was normalized to Renilla luciferase activity. Data are shown as means ± SEM (n = 3, *p<0.05 by Tukey’s test). (C) Degradation assay of CRY1 protein in HEK293 cells. Sixty hours after transfection, cells were treated with 100 μg/ml CHX and fractionated into the nuclear and cytosolic fractions. CRY1 protein levels at the starting point (t = 0 hr) were normalized to 1. Data are shown as means with SEM (n = 3*, p<0.05 by Student’s t-test).
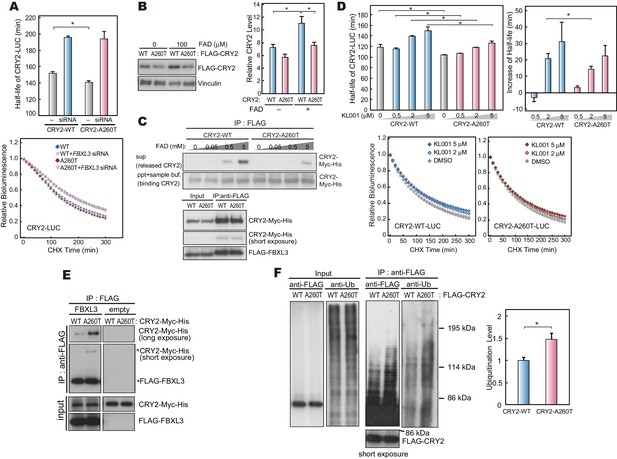
CRY2-A260T binds more strongly to FBXL3 leading to faster degradation of mutant CRY2.
(A) Effect of human FBXL3 knockdown on CRY2 protein stability in HEK293 cells. Forty-eight hours after transfection, the culture medium was changed to the recording medium containing 100 μg/ml CHX, and bioluminescence of CRY2-LUC was recorded continuously. Bioluminescence normalized to the value at time 0 was fitted to an exponential curve to determine half-life of CRY2-LUC. Data are shown as means with SEM (n = 4, *p<0.05 by Welch’s t-test). (B) Effect of FAD on CRY2 protein levels. Forty-two hours after transfection, HEK293 cells were treated with 100 μM FAD for 6 hr. Data are shown as means with SEM (n = 3, *p<0.05 by Student’s t-test). (C) FAD and FBXL3 competition assay. CRY2-FBXL3 complex expressed in HEK293 cells was purified using FLAG antibody. FAD was added to CRY2-FBXL3 complex and incubated at 4°C for 2 hr in vitro. (D) CRY2 protein stability in the cells treated with KL001. Twenty-four hours after transfection, the culture medium was changed to the recording medium containing 100 μg/ml CHX and KL001. Recording of bioluminescence and calculation of half-life was performed as described above. *p<0.05 by Student’s t-test (n = 3). (E) Interaction of CRY2 with FBXL3 in HEK293 cells treated with MG132 for 6 hr prior to harvesting. (F) Ubiquitylation of CRY2. HEK293 cells expressing FLAG-CRY2 were treated with MG132 for 6 hr before harvesting. FLAG-CRY2 was then purified and blotted with anti-Ubiquitin antibody. Quantitative data are shown as means with SEM (n = 3, *p<0.05 by Student’s t-test).
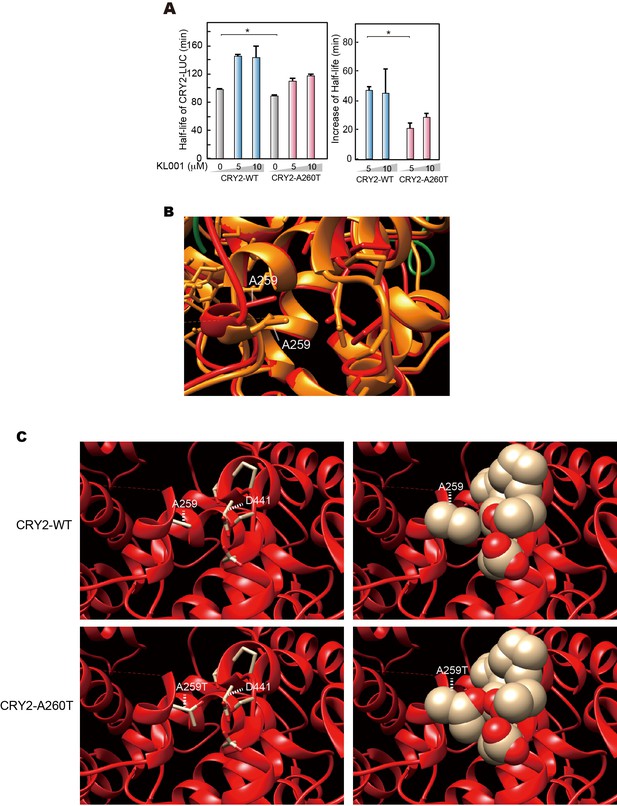
Structural modeling of A260T CRY2.
(A) CRY2 protein stability in the cells treated with KL001. Twenty-four hours after transfection, the culture medium was changed to the recording medium containing 100 μg/ml CHX and KL001 at the indicated concentration. *p<0.05 by Student’s t-test (n = 3). (B) Structure of mouse CRY2 bound to FAD (PDB code 4I6G) (orange) or FBXL3 (4I6J) (red). Green represents FBXL3. Short dashed lines indicate the side chain of A259 (A260 of human CRY2). CRY2 bound to FBXL3 is in a more open conformation compared to the FAD-bound CRY2. (C) Structure of mouse CRY2-WT (top) or mouse CRY2-A259T (corresponded to human CRY2-A260T, bottom) bound to FBXL3. The Ala to Thr mutation increases the molecular density and likely changes the conformation of CRY2 from the FAD-bound form to the FBXL3-bound form, in which main chain around A259 moved to left side in this image.
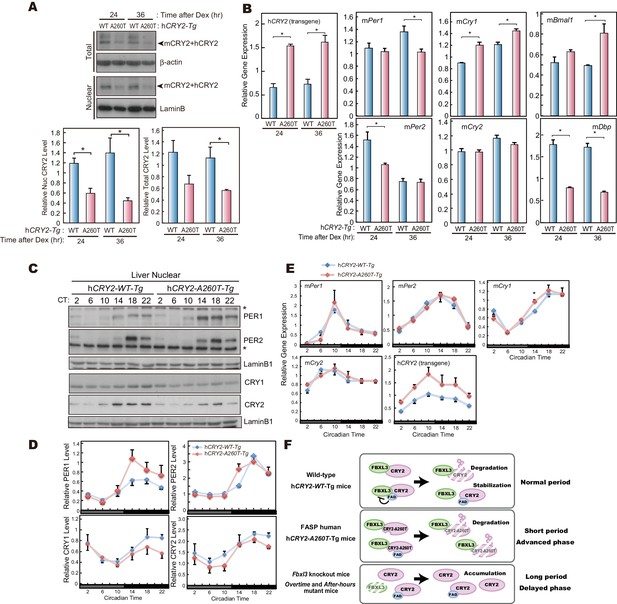
CRY2-A260T expression is down-regulated in hCRY2-A260T mice.
(A) CRY2 protein levels in synchronized MEFs. Cells were treated with 100 nM Dex for 2 hr to synchronize the cellular rhythms. Media was change and MEFs were cultured for 24 or 36 hr before harvesting. Quantified band intensities of CRY2 (mouse CRY2 and human CRY2) are shown as means ± SEM (n = 3, *p<0.05 by Student’s t-test). (B) mRNA levels of clock genes in synchronized MEFs. Cellular rhythms of MEFs were synchronized with 100 nM Dex for 2 hr. mRNA levels were quantified by real-time PCR. Data are shown as means ± SEM (n = 3, *p<0.05 by Student’s t-test). (C) Temporal expression profiles of PER1, PER2, CRY1 and CRY2 in mouse liver. Mice were sacrificed every 4 hr on the second day in DD. Asterisks mark non-specific bands. (D) Quantification of protein levels in (C). Data are shown as means ± SEM (n = 3). (E) mRNA levels of indicated clock genes in mouse liver. Mice were sacrificed every 4 hr on the second day in DD. mRNA levels of indicated genes were quantified by real-time PCR using gene specific primers. Data are shown as means ± SEM (n = 3). (F) Model of CRY2 protein regulation. In wild-type, FAD binding to CRY2 acts to stabilize by competing with FBXL3. In hCRY2-A260T transgenic mice or FASP human subjects with CRY2 mutations, FAD does not protect CRY2 from FBXL3-mediated degradation. Destabilization of CRY2 results in shortened period, leading to advanced sleep phase. In Fbxl3 knockout mice or mutant mice (Overtimeand After-hours) (Godinho et al., 2007; Siepka et al., 2007), CRY2 is stabilized in the nucleus, thus lengthening the circadian period.
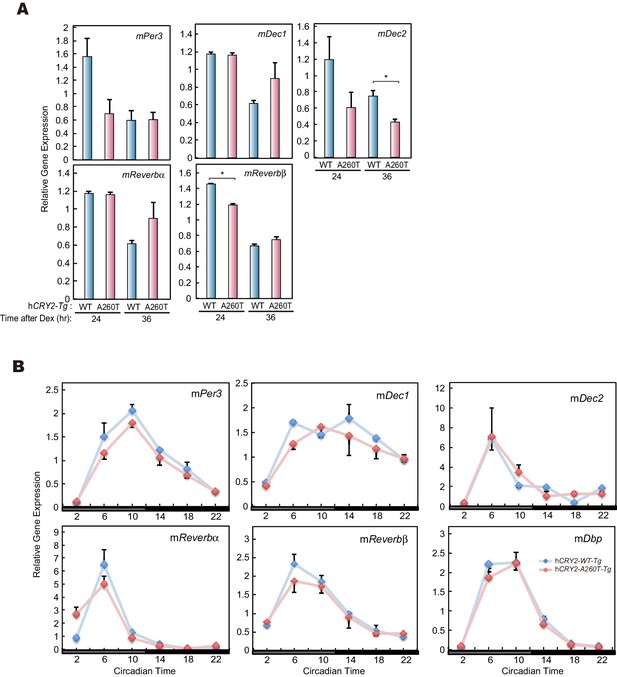
mRNA levels of clock genes in MEF and liver of hCRY2 BAC Tg mice.
(A) mRNA levels of indicated genes in synchronized MEFs. Cellular rhythms of MEFs were synchronized with 100 nM Dex for 2 hr. mRNA levels were quantified by real-time PCR. Data are shown as means ± SEM (n = 3, *p<0.05 by Student’s t-test). (B) Expression patterns of indicated clock genes in mouse liver. Mice were sacrificed every 4 hr on the second day in DD. mRNA levels of indicated genes were quantified by real-time PCR using gene specific primers. Data are shown as means ± SEM (n = 3).





