A filter at the entrance of the Golgi that selects vesicles according to size and bulk lipid composition
Figures
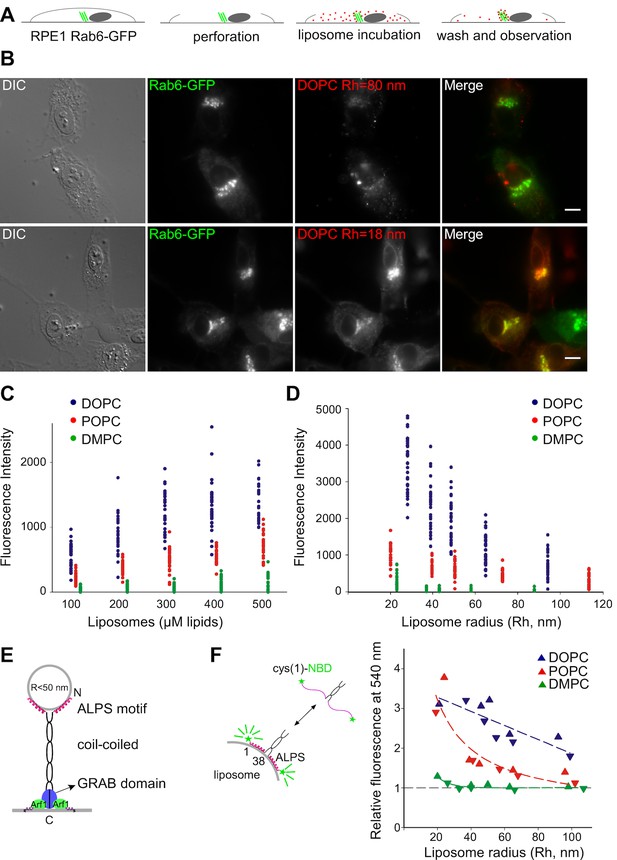
Preference of the Golgi for small unsaturated liposomes and biochemical properties of GMAP-210 ALPS motif.
(A) Principles of the Kobayashi-Pagano experiment. (B) Typical observations. RPE1 cells stably expressing Rab6-GFP were perforated and incubated with DOPC liposomes (labeled with 1 mol% Rhod-PE). The liposomes were prepared by sonication or by extrusion through 200 nm (diameter) filters. The actual liposome radius (Rh) is indicated. Images were acquired with an epifluorescence microscope. (C, D) Experiments similar to that shown in (B) with liposomes of defined composition (C) or size (D). In (C) all liposomes were obtained by extrusion through 30 nm filters. In (D) all liposomes were used at a concentration of 250 µM lipids. The liposome fluorescence intensity in the Golgi area was determined. Each point corresponds to one cell from two to three independent experiments. (E) Domain organization of GMAP-210 and minimal model for membrane tethering (Drin et al., 2008). (F) Effects of membrane curvature and lipid unsaturation on the binding of the N-ter region of GMAP-210 to liposomes. Liposome binding was determined by monitoring the NBD fluorescence intensity. The plot shows the effect of liposome size and unsaturation from two independent experiments (triangles and inverted triangles). The fluorescence in solution was set at 1. Bars: 10 µm.
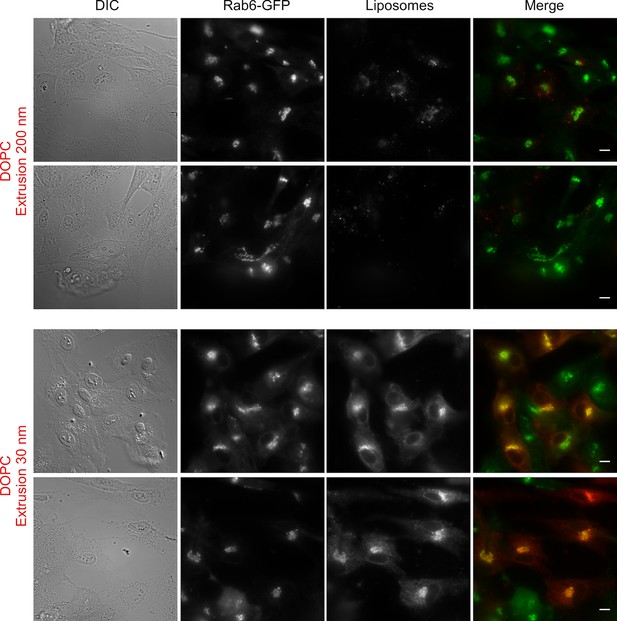
Liposome tethering properties of perforated cells.
Large fields of RPE1 cells stably expressing Rab6-GFP after perforation and incubation with DOPC liposomes (extrusion 200 nm or 30 nm). Scale bars: 10 µm.
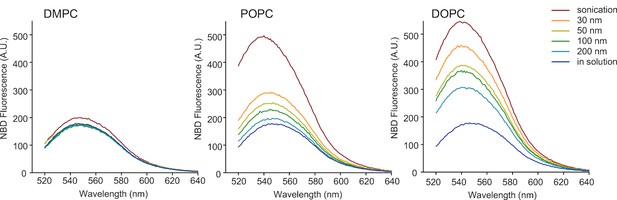
Cumulative effects of membrane curvature and lipid unsaturation on the binding of the N-ter region of GMAP-210 to liposomes.
Fluorescence spectra of mixtures containing 0.125 µM NBD-labeled GMAP-210 (fragment [1–375]) and liposomes (150 µM lipids) of the indicated size and composition. The fluorescence intensity at 540 nm was measured to build the plot of Figure 1F.
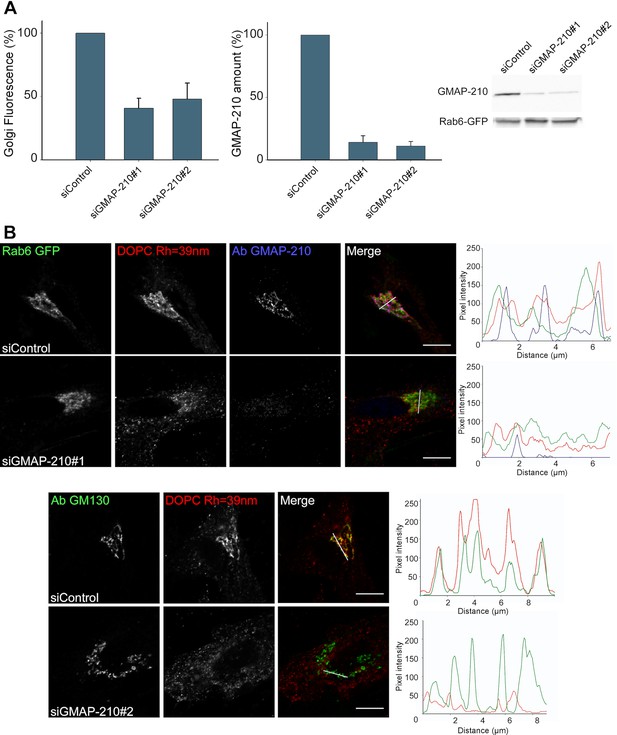
GMAP-210 is responsible for the ability of the Golgi to capture liposomes.
(A) RPE1 cells were treated with siRNA against GMAP-210 or with control siRNA. After 72 hr, the cells were perforated and small DOPC liposomes (250 µM lipids), which were obtained by extrusion through 30 nm polycarbonate filters, were applied. Liposome capture was quantified as in Figure 1. The western blot shows the amount of GMAP-210 in the siRNA-treated cells. (B) Immunofluorescence characterization of RPE1 cells after treatment with anti GMAP-210 siRNA or control siRNA followed by perforation and incubation with small DOPC liposomes obtained by extrusion. The actual liposome radius is indicated. The images show one Z section as obtained in a confocal microscope. The profiles show the fluorescence intensity of the three markers along the horizontal lines as indicated in the merged images. Bars: 10 µm. Error bars: SEM (n = 4)
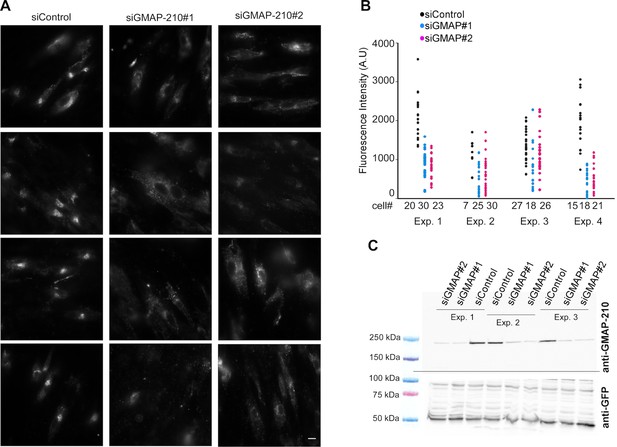
GMAP-210 is responsible for the ability of the Golgi to capture liposomes.
(A) RPE1 cells were treated with siRNA against GMAP-210 or with control siRNA. After 72 hr, the cells were perforated and small DOPC liposomes (250 µM lipids), which were obtained by extrusion through 30 nm polycarbonate filters, were applied. (B) Quantification of liposome capture from 4 independent experiments similar to (A). (C) Western blot analysis of the siRNA-treated cells. The data shown in (B) and (C) were used to build the bar plots shown in Figure 2A. Scale bars: 10 µm.
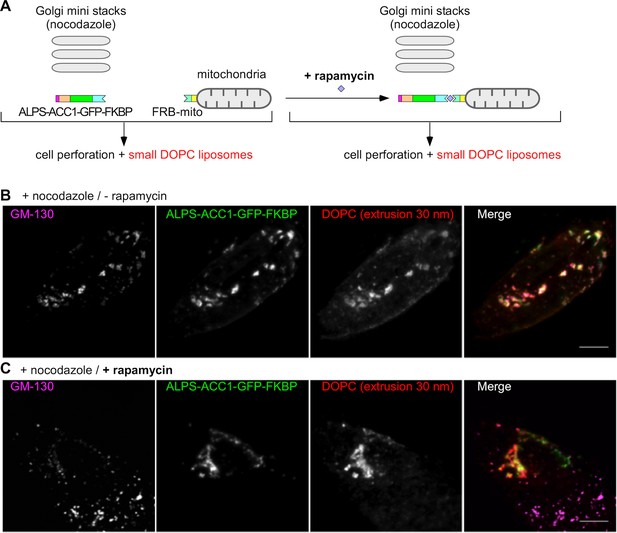
Mitochondria-anchored GMAP-210 ALPS competes with Golgi mini stacks for the capture of small liposomes.
(A) Scheme of the constructs and principle of the experiment. Cells expressing ALPS-ACC1-GFP-FKBP and Mito-FRB were treated with nocodazole to disperse the Golgi apparatus into mini stacks. Thereafter, the cells were either treated with rapamycin or not, perforated, and incubated with small DOPC liposomes (extrusion: 30 nm). (B) In the absence rapamycin, small liposomes (extrusion 30 nm) colocalize with ALPS-ACC1-GFP-FKBP and with the Golgi mini stacks (as stained by anti GM130 antibody). (C) In the presence rapamycin, the small liposomes and ALPS-ACC1-GFP-FKBP colocalize in many regions that are negative for GM130. All images were acquired with a confocal microscope and were analyzed using Volocity software. These experiments were repeated three times. Data show representative images. Scale bars: 10 µm.
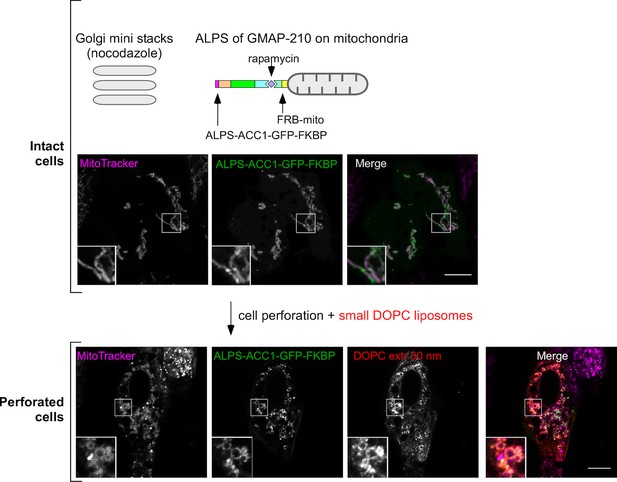
Competition between mitochondria-anchored GMAP-210 ALPS and Golgi mini stacks for liposome capture.
Cells were treated with nocodazole to disperse the Golgi apparatus into mini stacks and then with rapamycin to attach ALPS-ACC1-GFP-FKBP to mitochondria through Mito-FRB. Thereafter, the cells were perforated and incubated with small DOPC liposomes (extrusion: 30 nm). The mitochondria were visualized with MitoTracker. Images were acquired with a confocal microscope. Note that cell perforation causes the mitochondria network to break into small circular elements, around which ALPS-ACC1-GFP-FKBP and small DOPC liposomes were found. These experiments were repeated three times. Data show representative images. Scale bars: 10 µm.
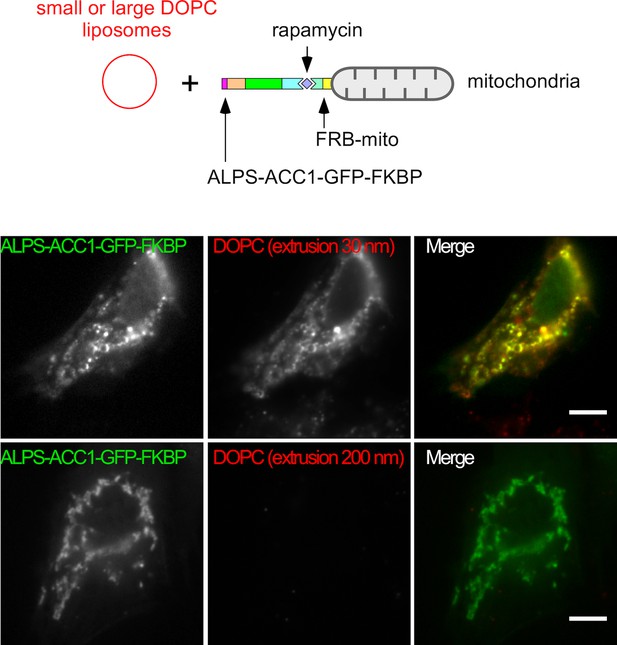
Mitochondria-anchored ALPS of GMAP-210 captures small but not large liposomes.
Cells were treated with nocodazole to disperse the Golgi apparatus into mini stacks and with rapamycin to attach ALPS-ACC1-GFP-FKBP to mitochondria through Mito-FRB. Thereafter, the cells were perforated and incubated with small (extrusion: 30 nm) or large (extrusion: 200 nm) DOPC liposomes. After several washes, images were acquired with a confocal microscope and were analyzed using Volocity software. Note that only small liposomes are captured by the ALPS construct. These experiments were repeated three times. Data show representative images. Scare bars: 10 µm.
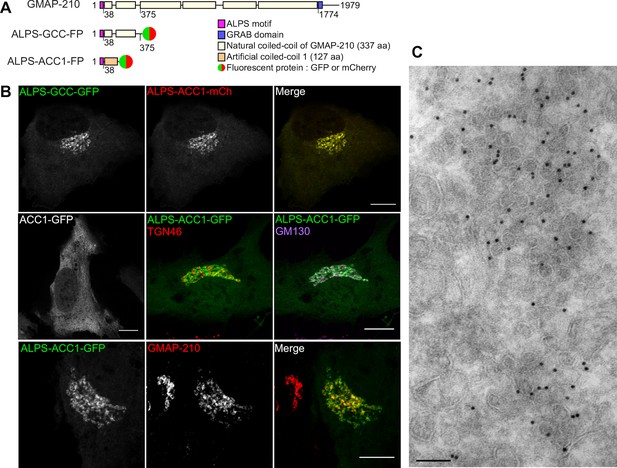
The ALPS motif of GMAP-210 targets cis Golgi vesicles.
(A) Scheme of the constructs. The first construct corresponds to the N-terminal region of GMAP-210 (aa 1–375; ALPS = 1–38) fused to a C-ter fluorophore (GFP or mCherry). Alternatively, the ALPS motif of GMAP-210 was introduced upstream of an artificial coiled-coil (ACC1). (B) Subcellular localization of GCC- and ACC-based constructs by confocal fluorescence microscopy. Golgi localization was preserved by replacing the coiled-coil region of GMAP-210 (GCC) by an artificial coiled-coil (ACC1), but was abolished by deletion of the ALPS motif. Immunofluorescence of cells expressing ALPS-ACC1-GFP shows low colocalization with TGN46, intermediate overlap with GM130, and high colocalization with endogenous GMAP-210. All images were acquired with a confocal microscope and were analyzed using Volocity software. Scale bars: 10 µm, Z plane. (C) Immunogold EM of RPE1 cells expressing ALPS-ACC1-GFP. Scale bars: 10 µm (B) 100 nm (C). These experiments were repeated at least three times, except EM, which was repeated twice. Data show representative images.
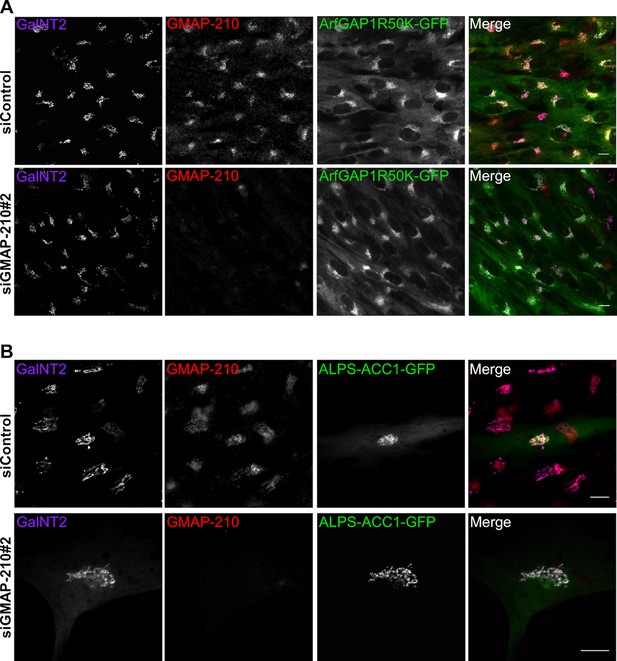
GMAP-210 depletion by siRNA does not affect the Golgi targeting of ALPS-containing probes.
(A) RPE1 cells stably expressing ArfGAP1(R50K mutant)-GFP were transfected with siGMAP-210#2 or control siRNA for 72 hr. (B) RPE1 cells were transfected with siGMAP-210#2 or with control siRNA for 72 hr and then with ALPS-ACC1-GFP for 7 hr. In A and B, cells were then fixed, immunostained with antibodies against GMAP-210 and the cis-Golgi marker GalNT2, and visualized by confocal microscopy. Z plane, scale bar: 10 µm.
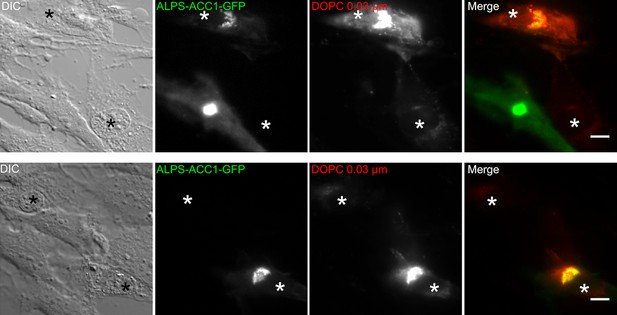
ALPS-ACC1-GFP increases the liposome capture properties of cells.
RPE1 cells were transitory transfected with ALPS-ACC1-GFP for 7 hr. Thereafter, the cells were perforated and 250 µM DOPC liposomes (extrusion 0.03 µm) were added for 5 min at 30°C. After several washes, images were acquired with a wide-field microscope and processed with ImageJ. Perforated cells are indicated by an asterisk. Those that express ALPS-ACC1-GFP show much higher liposome signal than those that do not express the construct. Scale bar: 10 µm.
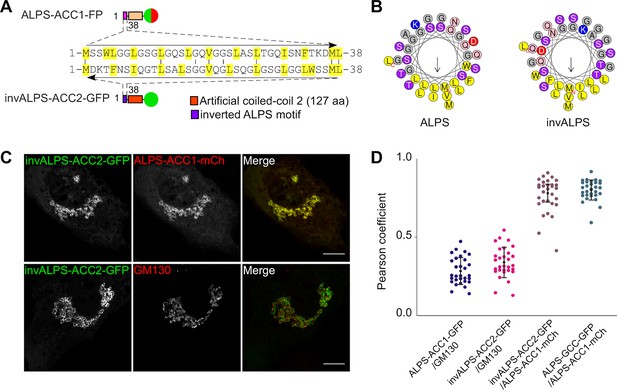
Selective targeting of the ALPS motif of GMAP-210 to cis Golgi vesicles without stereospecific interactions.
(A, B) Sequence comparison of the ALPS motif of GMAP-210 and the corresponding inverted motif (invALPS). The two sequences have low identity (≈ 25%; A), but helical wheel-representations (B) show that they display the same amphipathic character. Helical representations were made with Heliquest (Gautier et al., 2008).(C) A coiled-coil construct harboring an inverted ALPS motif (invALPS-ACC2-GFP) displays the same subcellular localization as a construct harboring the original ALPS motif (ALPS-ACC1-mCherry). (D) Pearson coefficient between the green and red channels for the constructs shown in (C) as well as for other combinations of constructs or endogenous proteins. 30 cells were examined from 3 independent experiments. The vertical black bars show the mean ± SD. All images were acquired with a confocal microscope and were analyzed using Volocity software. Scale bars: 10 µm, Z plane.
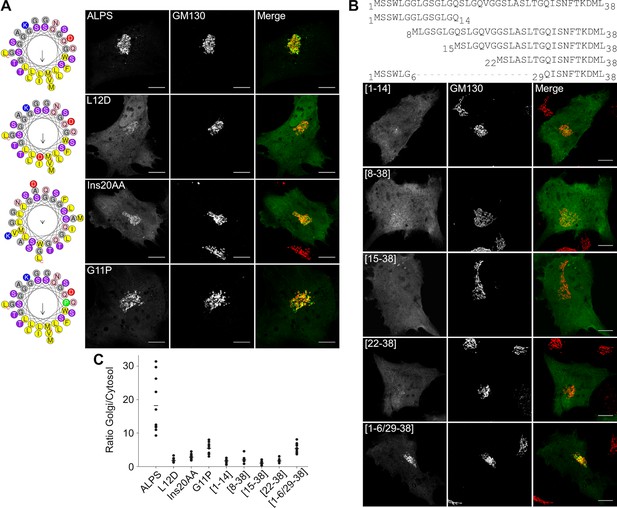
The ALPS motif of GMAP-210 interacts with Golgi vesicles as an amphipathic helix.
(A) Effect of mutations disrupting the amphipathic character of the ALPS motif of GMAP-210. For each mutant, a helical plot made with Heliquest (Gautier et al., 2008) is shown. Mutation L12D introduces a negative charge in the hydrophobic face. Ins20AA corresponds to 2 alanines inserted in the middle of the ALPS sequence. G11P should rigidify the sequence. (B) Effect of gradual truncation of the ALPS sequence. All mutants shown in (A) and (B) were derived from ALPS-ACC1-GFP. (C) Quantification. For both (A) and (B), ten cells were analyzed for each mutant. Once the background was subtracted, two ROI with same area were applied in the Golgi and in the cytosol. The average fluorescence was determined in each ROI and the Golgi/cytosol ratio was then calculated. All images were acquired with a confocal microscope and were analyzed using Volocity software. Scale bars: 10 µm, Z projections. Scale bars: 10 µm. The figure shows one experiment where all constructed were transfected in parallel and ten cells were analyzed for each condition. The horizontal bars show the means. Other experiments were performed with subgroups of mutants and gave similar results.
Effects of point mutations on the Golgi localization of the ALPS motif of GMAP-210.
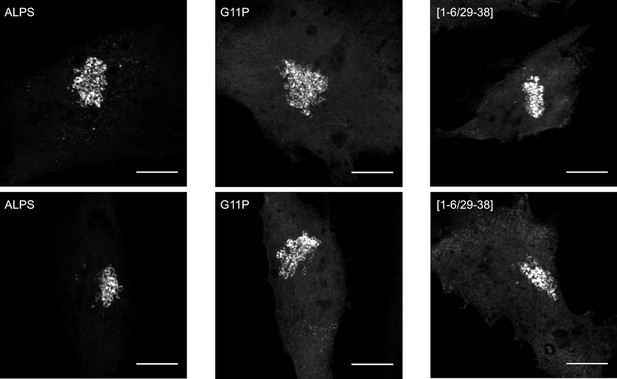
This figure shows different images of the subcellular localization of some mutants described in Figure 6. The mutants G11P and [1–6/29–38] have a clear Golgi localization. However, their cytosolic fluorescent signal is much higher than that of wild-type ALPS-ACC1-GFP, implying a lower Golgi/cytosol ratio (see Figure 6C). Scale bar: 10 µm.
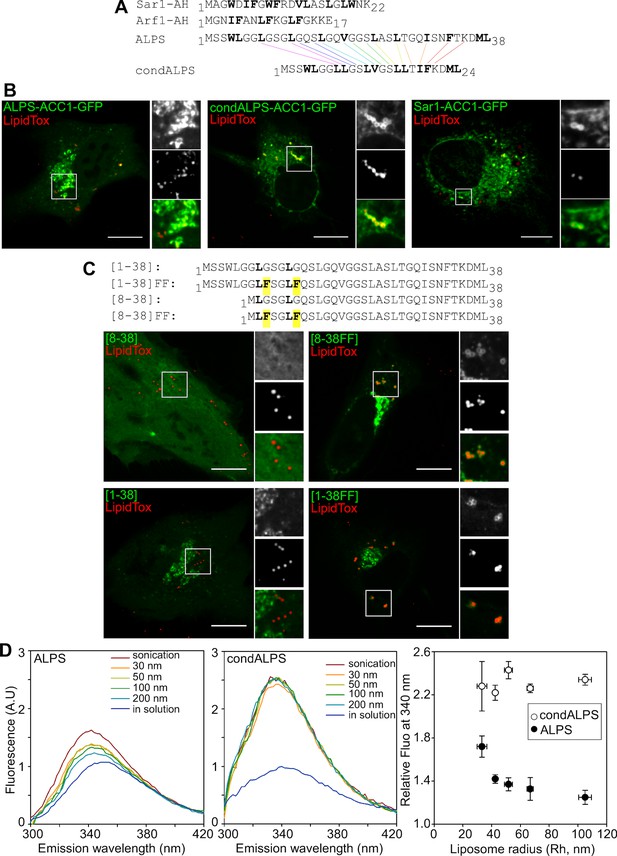
The sparse distribution of hydrophobic residue in ALPS determines its selective cellular targeting and its sensitivity to membrane curvature.
(A) Sequence comparison between the ALPS motif of GMAP-210 and the amphipathic helices of Arf1 and Sar1. Hydrophobic residues are shown in bold. In ALPS, most hydrophobic residues are sparse. In Arf1 and Sar, most hydrophobic residues are paired. By removing polar residues between sparse hydrophobic residues, the ALPS sequence was condensed (condALPS) to create hydrophobic pairs. (B) Localization of ALPS-ACC1-GFP, condALPS-ACC1-GFP, and Sar1-ACC1-GFP in RPE1 cells. Lipid droplets were revealed by lipidTox staining (red). Imaged were acquired from living cells using a spinning disk microscope. Z planes, scale bar: 10 µm. (C) Effect of adding hydrophobic residues in the ALPS 1–38 sequence or in the 8–30 truncated mutant. All mutants derived from the ALPS-ACC1-GFP construct. (D) Sensitivity of ALPS and condALPS peptides to membrane curvature. The peptides (1 µM) were incubated with liposomes (lipids 0.5 mM) of defined size. Tryptophan fluorescence was measured between 300 to 450 nm. The right plot shows the relative fluorescence at 340 nm as a function of liposome radius, setting the fluorescence in solution at 1. Error bars: SEM from four independent experiments. Scale bars: 10 µm.
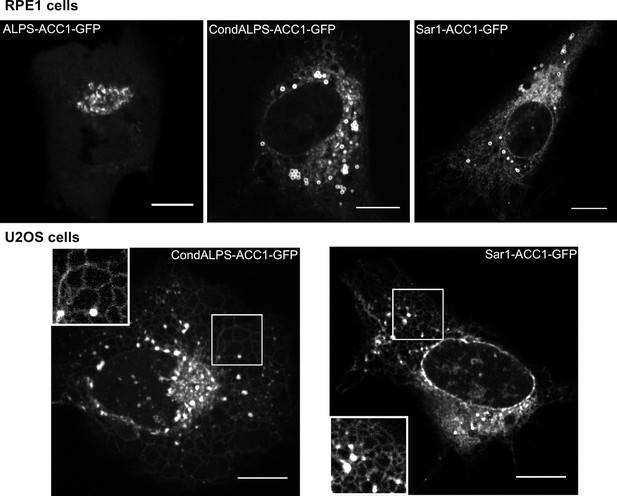
Subcellular localization of ALPS-ACC1-GFP, condALPS-ACC1-GFP, and Sar1-ACC1-GFP in RPE1 and U2OS cells.
In RPE1 cells, condALPS-ACC1-GFP and Sar1-ACC1-GFP intensively labeled lipid droplets and to a lesser extent the ER and the nuclear envelope. In U2OS cells, the ER localization of condALPS-ACC1-GFP and Sar1-ACC1-GFP is more obvious given the reticular pattern of the ER in these cells. Imaged were acquired from living cells using a spinning disk microscope. Z planes, scale bar: 10 µm.
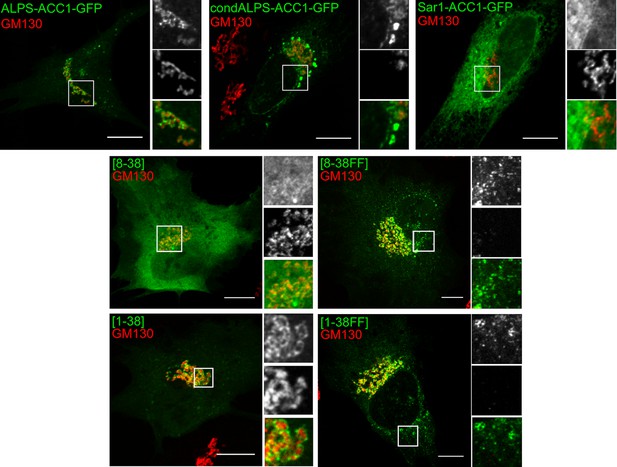
The sparse distribution of hydrophobic residue in ALPS determines its selective cellular targeting.
This figure shows the subcellular localization of the same constructs as shown in Figure 7 but in comparison with the cis Golgi marker GM130. Whereas wild-type ALPS displays Golgi localization, mutants with hydrophobic pairs bind to other organelles besides the Golgi (e.g. nuclear envelope, ER network, lipid droplets). Scale bar: 10 µm.
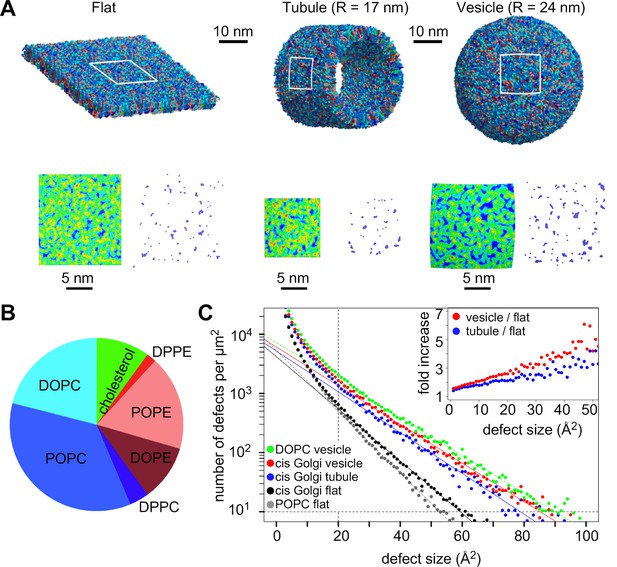
Distribution of lipid packing defects in model cis Golgi membranes.
(A) Typical coarse grained models of lipid bilayers with a lipid composition similar to that of COPI vesicles (Brügger et al., 2000) and with a flat, tubular or spherical geometry. (B) Lipid composition of the bilayers. (C) Lipid packing defect distribution. For comparison, the results from simulations with flat POPC or spherical DOPC membranes (Vanni et al., 2014) are also shown. The inset shows the ratio between the number of defects in the vesicle or in the tube as compared to the flat membrane.





