POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in Arabidopsis
Figures

HDA9 interacts with PWR and WRKY53.
(A) Summary of partial proteins associated with HDA9 and PWR identified by affinity purification and mass spectrometry analysis. Coverage indicates the percentage of full-length protein covered by identified unique peptides. Unique peptides indicate the number of identified peptides that are mapped to an individual protein. (B) Co-immunoprecipitation of HDA9 and PWR using Arabidopsis F1 plants expressing both HDA9-HA and PWR-FLAG. (C) In vitro pull down assay of GST-WRKY53 and HDA9-FLAG. Two technical replicates were performed (rep1 and rep2). GST protein serves as a control.
-
Figure 1—source data 1
List of proteins identified by IP-MS in HDA9 and PWR.
- https://doi.org/10.7554/eLife.17214.004

HDA9-FLAG protein is functional in Arabidopsis.
(A) Determination of HDA9 protein level in transgenic Arabidopsis expressing pHDA9::HDA9-3xFLAG/hda9 (HDA9-FLAG). Top panel, schematic diagram of HDA9-FLAG protein; bottom panel, detection of HDA9-FLAG protein with FLAG antibody. Coomassie staining of large subunit of rubisco as a loading control. (B) HDA9-FLAG rescued the dwarf phenotype of hda9 mutant.
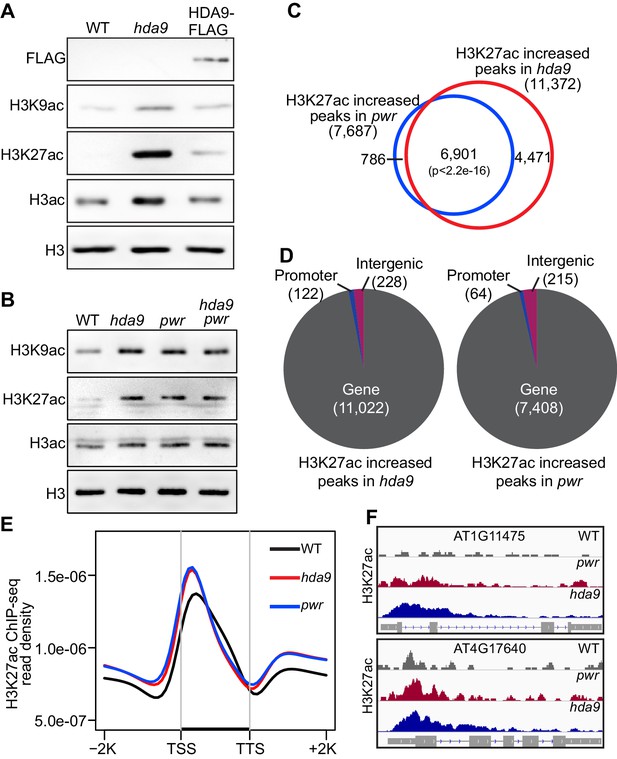
Loss-of-function hda9 and pwr mutants induce H3K9 and H3K27 hyperacetylation.
(A) Immunoblots of histone acetylation marks in hda9, pwr, and HDA9-FLAG early senescent leaves. (B) Immunoblots of histone acetylation marks in hda9, pwr, and hda9 pwr early senescent leaves. (C) Overlap of H3K27ac increased peaks in hda9 and pwr identified by ChIP-seq. Fisher’s exact test was used to calculate the p-value. (D) Genomic distribution of H3K27ac increased peaks in hda9 and pwr. (E) Metaplots of the H3K27ac distribution on genes in WT, hda9, and pwr. Black bar in the X-axis represents all genes in the genome. TSS, Transcription Start Sites; TTS, Transcription Terminal Sites; −2K and +2K represent 2 kb upstream of TSS and 2 kb downstream of TTS, respectively. The Y-axis represents read density of H3K27ac ChIP-seq. (F) Browser snapshots of representative loci with increased H3K27ac in hda9 and pwr.
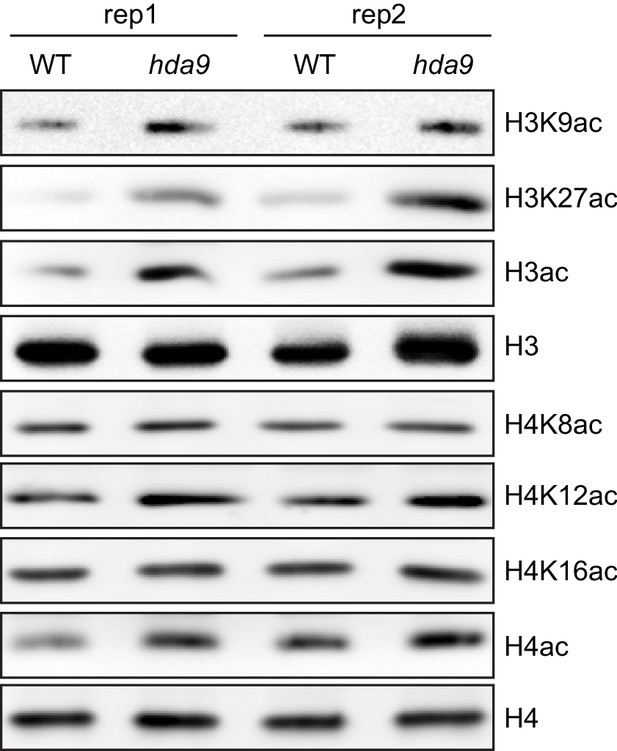
HDA9 preferentially removes acetylation on histone H3 tail in vivo.
Two biological replicates (rep1 and rep2) were performed.
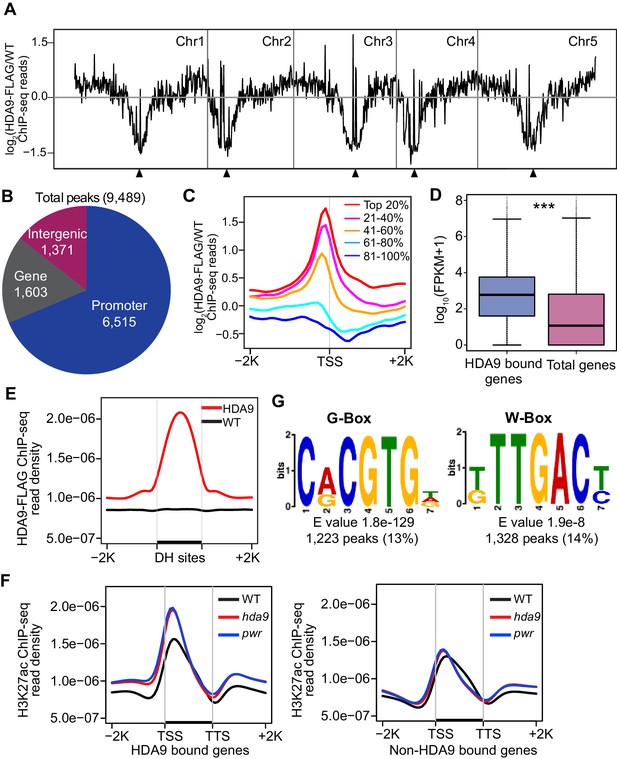
HDA9 binds to promoters of active genes.
(A) Chromosomal views of HDA9 distribution on five chromosomes. The Y-axis represents the log2 value of HDA9-FLAG ChIP-seq reads relative to those of untagged WT control. Chr1, Chr2, Chr3, Chr4, and Chr5 represent chromosomes 1 to 5, respectively. Black triangles indicate the location of centromeric regions. (B) Genomic distribution of HDA9 binding peaks. (C) Metaplots of HDA9 binding levels on genes. Total genes were divided evenly into five groups based on their expression level in WT. Top 20% indicates the 20% genes with highest expression level, 81%–100% indicates the 20% genes with lowest expression level. The Y-axis represents the log2 value of HDA9-FLAG ChIP-seq reads relative to those of untagged WT control. −2K and +2K represent 2 kb upstream and downstream of TSS, respectively. (D) Box plots of the average expression levels of HDA9 bound genes and total genes. The Y-axis indicates log10 value of FPKM + 1. FPKM, Fragments Per Kilobase of transcript per million mapped reads. Bars within the boxes represent the mean values. ***p<0.001. (E) Metaplots of HDA9 binding on previously identified DH (DNase I Hypersensitive) sites in HDA9-FLAG and untagged WT control. Black bar in the X-axis represents DH sites. The Y-axis represents the read density of HDA9-FLAG ChIP-seq reads. (F) Metaplots of H3K27ac on HDA9 bound genes and non-HDA9 bound genes in WT, hda9, and pwr. Black bar in the X-axis represents genes. The Y-axis represents read density of H3K27ac ChIP-seq reads. (G) Representative DNA motifs identified in HDA9 binding sites by DREME.
-
Figure 3—source data 1
HDA9 binds to active genes.
(A) List of HDA9 bound genes. (B) Genes showing HDA9 binding and upregulation in hda9. (C) Chromatin related proteins showing HDA9 binding.
- https://doi.org/10.7554/eLife.17214.009
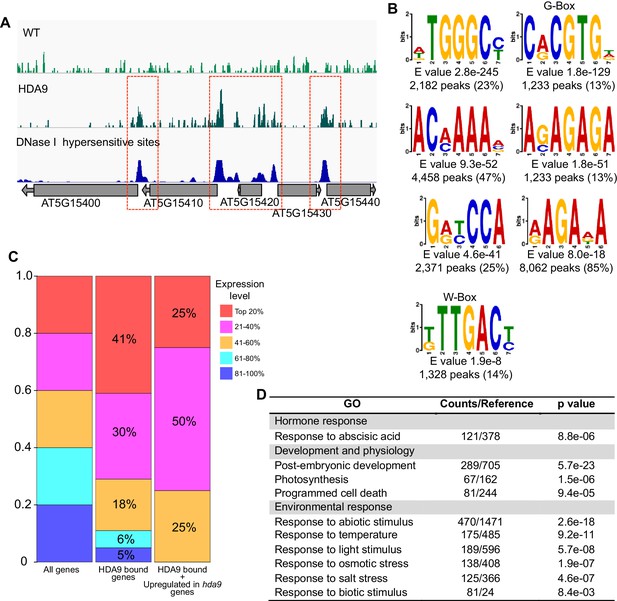
HDA9 binds to open chromatin regions with known DNA motifs.
(A) Representative snapshots show the correlations between HDA9 binding sites and DNase I hypersensitive sites. (B) DNA motifs identified in HDA9 binding peaks by DREME. Motifs with E value less than 1.0e-7, peak number more than 1200 were shown. E value and peak numbers are listed at bottom of each motif. (C) Analysis of expression level of HDA9 bound genes. Left panel, all genes are equally divided into 5 groups based on their expression level. Middle panel, distribution of HDA9 bound genes in different groups. Right panel, distribution of genes with HDA9 binding and upregulation in hda9 in different groups. (D) GO analysis of genes with HDA9 binding but no significant expression change in hda9.
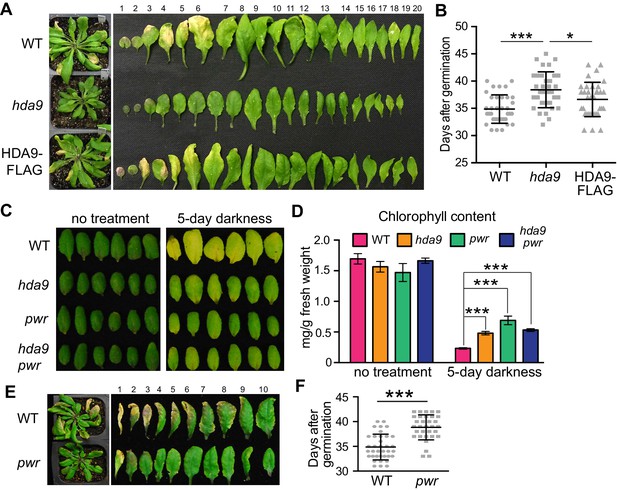
HDA9 and PWR act in the same pathway to promote leaf senescence.
(A) Phenotypic analysis of leaves from 5-week-old plants of wild type (WT), hda9, and HDA9-FLAG plants expressing HDA9-FLAG driven by the native HDA9 promoter in hda9 mutant background. Rosette leaves were numbered from bottom to top with the first leaf being the oldest and 20th being the youngest. (B) Quantification of days from germination to onset of leaf senescence in WT, hda9 and HDA9-FLAG. Points (round, square, or triangle) represent the number of days for an individual plant to reach onset of senescence. Error bars represent standard deviation for at least 30 tested plants. (C) Dark treatment of the 5th and 6th leaves detached from 3-week-old plants of hda9, pwr, and hda9 pwr double mutants. (D) Chlorophyll content measurement of leaves from (C). Error bars represent a standard deviation for three biological replicates. (E) Leaf senescence phenotype of 5-week-old pwr mutant. The oldest ten leaves are shown. (F) Quantification of days from germination to the onset of leaf senescence in WT and pwr. Points (round or square) represent the number of days for an individual plant to reach onset of senescence. Error bars represent standard deviations for at least 30 tested plants. Student’s t-tests were used to calculate the p values. *p<0.05, ***p<0.001.
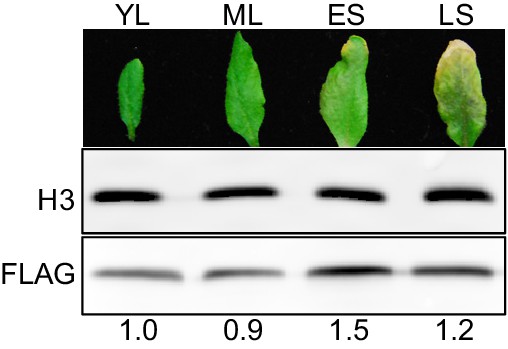
HDA9-FLAG shows elevated protein accumulation in early senescent leaves.
Western blots of HDA9-FLAG protein levels in young leaf (YL), mature leaf (ML), early senescent leaf (ES), and late senescent leaf (LS). Numbers at the bottom represent the intensity of HDA9-FLAG signals normalized to those of histone H3.
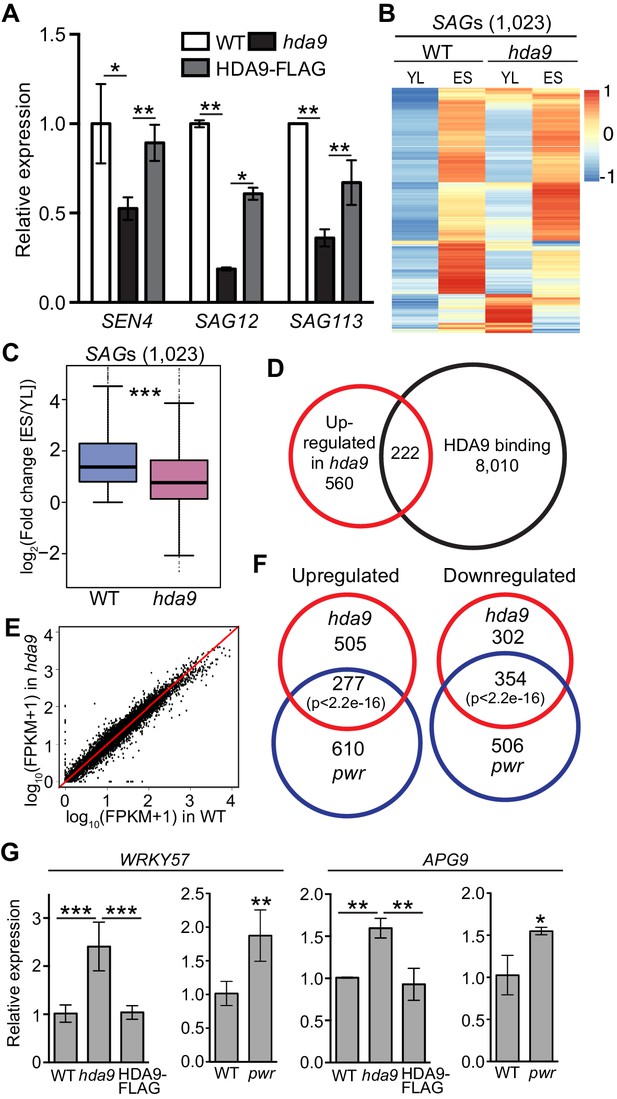
HDA9 and PWR regulate expression of the same group of genes involved in leaf senescence.
(A) Expression of senescence marker genes in hda9 by quantitative RT-PCR. Error bars represent a standard deviation from two biological replicates. (B) Heatmaps show expression of senescence-associated genes (SAGs) in young leaf (YL) and early senescence leaf (ES) in WT and hda9. The color bar on the right indicates the Z-score. (C) Boxplots show the less increased expression of SAGs in hda9 than WT in ES. The Y-axis represents log2 value of fold change of expression levels of SAGs between ES and YL. (D) Overlap of upregulated genes in hda9 and HDA9 bound genes. (E) Scatter plots show the expression of HDA9 bound genes in WT and hda9. (F) Overlap of differentially expressed genes in RNA-seq of hda9 and pwr. Fisher’s exact test was used to calculate the p-value. (G) Quantitative RT-PCR confirming the upregulation of WRKY57 and APG9 in hda9 and pwr. Relative expression was calculated as relative to ACTIN7, and then normalized to WT. Error bars represent a standard deviation from two biological replicates. *p<0.05, **p<0.01, ***p<0.001.
-
Figure 5—source data 1
Differentially expressed genes in hda9.
- https://doi.org/10.7554/eLife.17214.014
-
Figure 5—source data 2
Differentially expressed genes in pwr.
- https://doi.org/10.7554/eLife.17214.015
-
Figure 5—source data 3
List of overlapped genes showing differential expression in both hda9 and pwr.
- https://doi.org/10.7554/eLife.17214.016

HDA9 and PWR regulate similar group of genes involved in leaf senescence.
(A) Heatmaps show expression of senescence downregulated genes (SDGs) in young leaf (YL) and early senescence leaf (ES) in WT and hda9. The color bar on the right indicates the Z-score. (B) Boxplots show the less decreased expression of SDGs in hda9 than WT in ES. The Y-axis represents log2value of fold change of expression levels of SDGs between ES and YL. Bars within the boxes represent the mean values. ***p<0.001. (C) Heatmaps show expression of selected ABA responsive genes in YL and ES in WT and hda9. The color bar on the left indicates the Z-score. (D) A volcano plot of differentially expressed genes in ES in hda9. The Y-axis represents log10 of p value and the X-axis represents log2value of fold change in hda9 compared to WT. Genes were considered significantly downregulated or upregulated if p<0.05. (E) GO analysis of differentially expressed genes in hda9. (F) Venn diagram of genes with HDA9 binding and upregulated in hda9 and genes with increased H3K27ac in hda9. (G) Schematic symbols of candidate genes for molecular analysis selected from HDA9 bound genes with upregulation in hda9. ΔAlthough the increased expression of WRKY57 is not defined as significant (p<0.05) in hda9 (p=0.07), we confirmed the upregulation with RT-qPCR in both hda9 and pwr. (H) A volcano plot of differentially expressed genes in ES in pwr. The Y-axis represents log10 of p value and the X-axis represents log2value of fold change in pwr compared to WT. Genes were considered significantly downregulated or upregulated if p<0.05. (I) GO analysis of upregulated and downregulated genes in both hda9 and pwr.
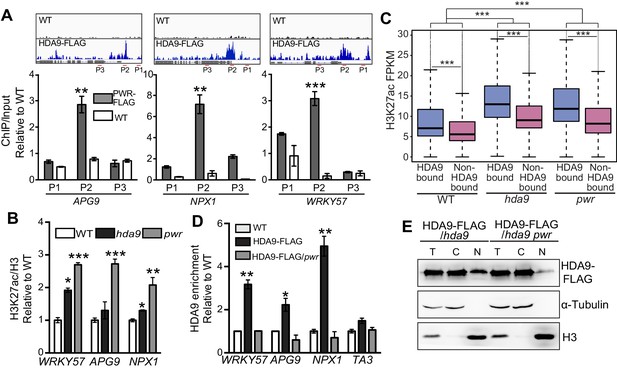
HDA9 nuclear accumulation and chromatin association are dependent on PWR.
(A) ChIP-qPCR shows that PWR is enriched at the same genomic regions of HDA9 targets. Upper panel illustrates snapshots of HDA9 binding at APG, NPX1, and WRKY57. ChIP-qPCR value of PWR was normalized to WT control. Primer positions are indicated with P1, P2, and P3. Error bars represent a standard deviation from two biological replicates. (B) ChIP-qPCR shows H3K27ac levels at WRKY57, APG9, and NPX1 in WT, hda9, and pwr mutants. ChIP-qPCR value of H3K27ac was normalized to WT control. Error bars represent a standard deviation from two biological replicates. (C) Box plots of H3K27ac levels on HDA9 bound genes and non-HDA9 bound genes in WT, hda9, and pwr. The Y-axis represents FPKM of H3K27ac ChIP-seq reads. Student’s t test, ***p<0.001. (D) ChIP-qPCR shows HDA9 enrichment on WRKY57, APG9, and NPX1 in HDA9-FLAG and HDA9-FLAG/pwr plants. TA3 is a transposable element that serves as a negative control. Error bars represent a standard deviation from two biological replicates. *p<0.05, **p<0.01. (E) Detection of HDA9-FLAG protein in total (T), cytoplasmic (C), and nuclear (N) extracts in HDA9-FLAG/hda9 and HDA9-FLAG/hda9 pwr.
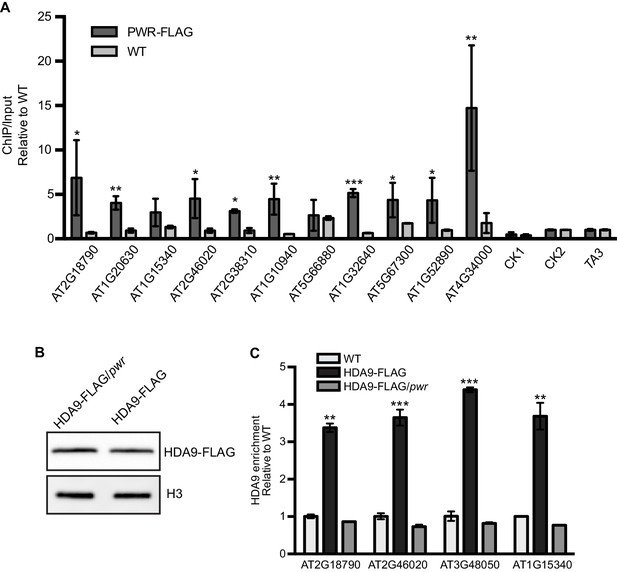
PWR is required for HDA9 recruitment to targets.
(A) ChIP-qPCR for PWR enrichment on randomly chosen HDA9 targets. CK1 (AT1G70380), CK2 (AT2G31425), and TA3 (AT1TE46405) are negative controls without HDA9 binding. Error bars represent a standard deviation from two biological replicates. (B) Western blot of HDA9-FLAG protein in WT and pwr background. (C) ChIP-qPCR for HDA9 enrichment on randomly chosen HDA9 targets in HDA9-FLAG/hda9 and HDA9-FLAG/hda9 pwr. Error bars represent a standard deviation for two biological replicates. Student’s t-test, *p<0.05, **p<0.01, ***p<0.001.
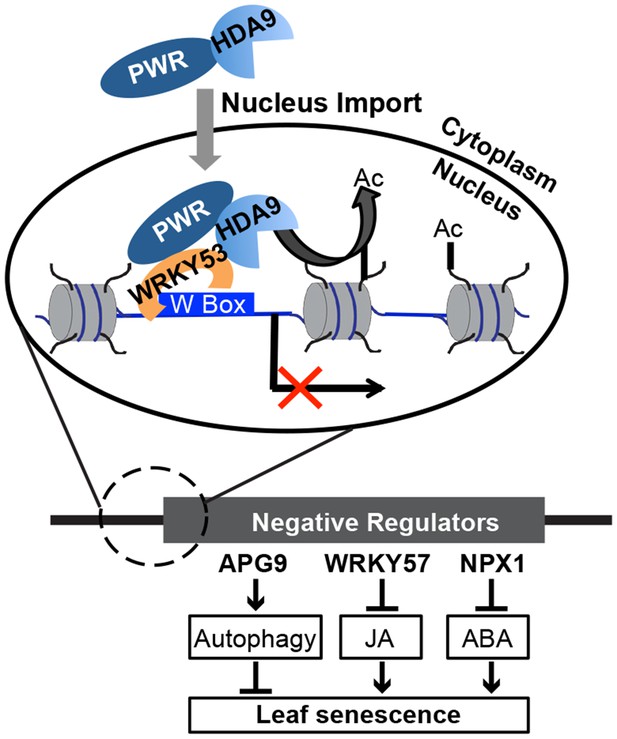
A working model for the biological functions and molecular mechanisms of HDA9 and PWR.
In the cytoplasm, PWR forms complexes with HDA9 and is required for the transport of HDA9 from the cytoplasm into the nucleus. In the nucleus, PWR recruits HDA9 to W-box containing promoter regions likely with the help of WRKY53. HDA9 catalyzes the removal of H3 acetylation marks and suppresses the expression of negative senescence regulators (e.g. WRKY57, APG9, and NPX1), which in turn induces the derepression of their downstream target genes to promote leaf senescence.
Additional files
-
Supplementary file 1
Primers used in this study.
- https://doi.org/10.7554/eLife.17214.021
-
Supplementary file 2
Reads of RNA-Seq and ChIP-Seq.
- https://doi.org/10.7554/eLife.17214.022





