PTRF/Cavin-1 promotes efficient ribosomal RNA transcription in response to metabolic challenges
Figures
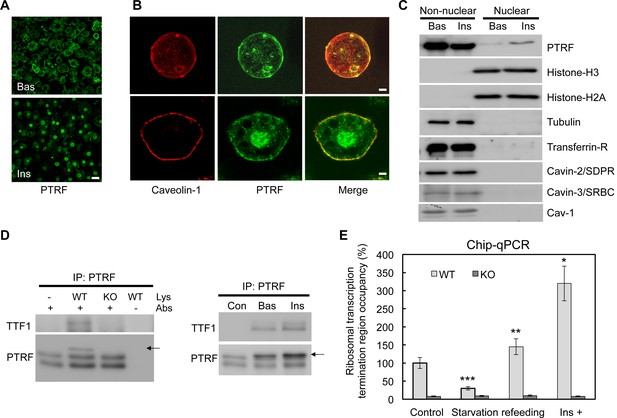
A functional role of PTRF in nucleus.
(A) Immunofluorescence staining of PTRF in basal and insulin stimulated 3T3-L1 adipocytes. Scale bars, 50 μm. (B) Higher magnificent images of insulin stimulated single isolated primary (top) and 3T3-L1 adipocyte (bottom) are shown, green: PTRF, and red: caveolin-1. Scale bars, 10 μm. (C) Nuclear and non-nuclear fractions from basal or stimulated primary adipocytes were subject to western blots by indicated antibodies. (D) Co-immunoprecipitation by PTRF antibody from basal and stimulated adipocytes nuclear lysate followed by western blots using TTF1 (Transcription Termination Factor, RNA Polymerase I) and PTRF antibodies, PTRF null adipocytes (KO) serving as a negative control for the co-IP. (E) Chip-qPCR assay using PTRF antibody from wild type control (WT) and PTRF null adipocytes (KO). *p<0.05, **p<0.01, and ***p<0.001; Student’s test. Error bars indicate SD.
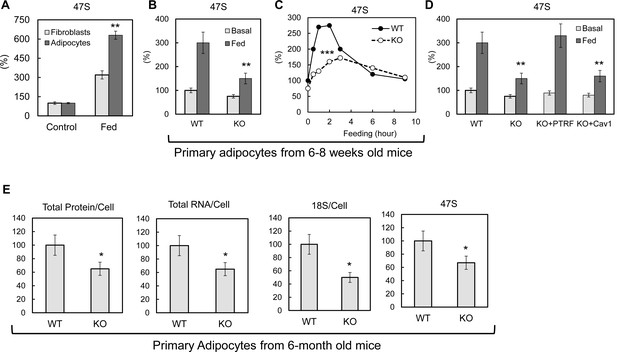
PTRF mediated ribosomal transcription regulations.
(A) Relative pre-rRNA (47S) levels were detected by RT-qPCR from 3T3-L1 fibroblast and fully differentiated adipocytes stimulated by switching culture medium from 1% BSA in PBS to full growth medium with insulin (Fed). (B) Basal (KRP buffer with 1% BSA) or 45 min stimulated (Fed: high glucose DMEM, 10% FBS, and insulin) pre-rRNA levels were detected by RT-qPCR in primary adipocytes isolated from wild type (WT) and PTRF null (KO) mice. (C) A stimulation time-course of B. (D) Pre-rRNA levels were detected by RT-qPCR from basal or stimulated wild type (WT), PTRF null (KO), PTRF re-transfected PTRF null (KO+PTRF), and Cav-1 re-transfected PTRF null (KO+Cav1) MEFs cells. (E) Relative levels of total proteins, RNA, 18S, and 47S from wild type or PTRF null mice (n = 6) were measured and normalized to cell numbers. *p<0.05, **p<0.01, and ***p<0.001; Student’s test. Error bars indicate SD.
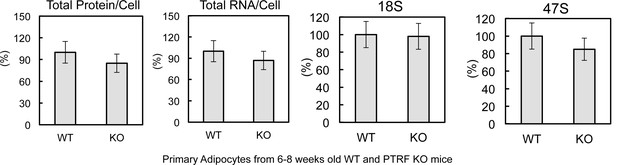
Characterization of ribosome biogenesis in 6–8 weeks old PTRF null and control mice.
Total proteins, RNAs, 18S, and 47S levels were measured from 6–8 weeks old wild type control and PTRF null mice (n = 6).
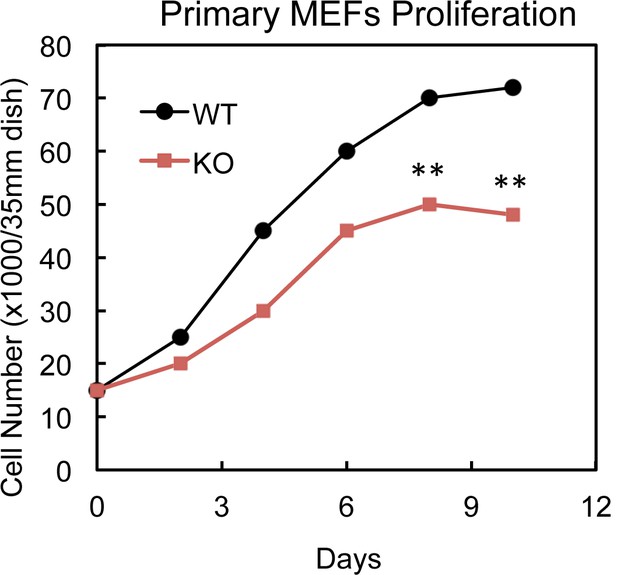
Cell growth curve of cultured WT and PTRF null MEFs cells.
Primary PTRF null (KO) and wild type control (WT) MEFs cell numbers were determined by cell counting on indicated days. **p<0.01; Student’s test. Error bars indicate SD.
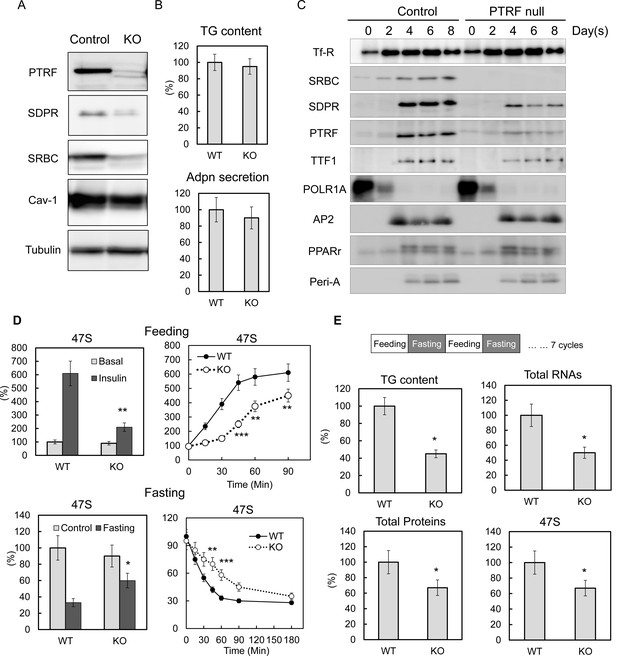
PTRF plays critical role mediating ribosomal transcription response to metabolic challenges in CRISPR/Cas9 genomic editing cell model, without any affect on differentiation.
(A) Indicated protein levels were detected by Western blots from CRISPR/Cas9 genomic edited PTRF null and control 3T3-L1 adipocytes. (B) Relative total TG content and adiponectin secretion levels were measured from CRISPR/Cas9 genomic edited PTRF null and control 3T3-L1 adipocytes. (C) Expression levels changes of indicated proteins during 3T3-L1 differentiation were measured by western blot from CRISPR/Cas9 genomic edited PTRF null and control 3T3-L1 adipocytes. (D) Pre-rRNA levels from nutrients/insulin stimulated (feeding: switching from PBS with 1% BSA to high-glucose DMEM with 10% FBS and insulin) or starved (Fasting: switching the growth medium to PBS with 1% BSA) were measure by RT-qPCR from CRISPR/Cas9 genomic edited PTRF null and control 3T3-L1 adipocytes. (E) After CRISPR/Cas9 genomic edited PTRF null and control 3T3-L1 adipocytes were subject to 'feeding' 12 hr followed by 'fasting' 12 hr cycle for seven days, total TG content, proteins, RNAs and 47S levels were measure as described before. *p<0.05, **p<0.01, and ***p<0.001; Student’s test. Error bars indicate SD.
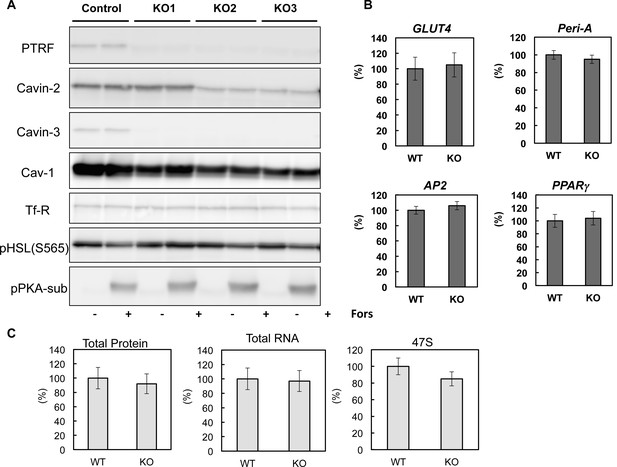
Characterizations of CRISPR/Cas9 genomic edited PTRF null 3T3-L1 adipocytes.
(A) Whole cell lysates from control and 3 cell lines of CRISPR/Cas9 genomic edited PTRF null 3T3-L1 adipocytes were separated on SDS-PAGE followed by immuno-blots using indicated antibodies. (B) Gene expression levels of some adipocyte specific markers were measured from control and one cell line (KO3) of CRISPR/Cas9 genomic edited PTRF null 3T3-L1 adipocytes by RT-qPCR. (C) Total proteins, RNAs and 47S were measured from control and one cell line (KO3) of CRISPR/Cas9 genomic edited PTRF null 3T3-L1 adipocytes.
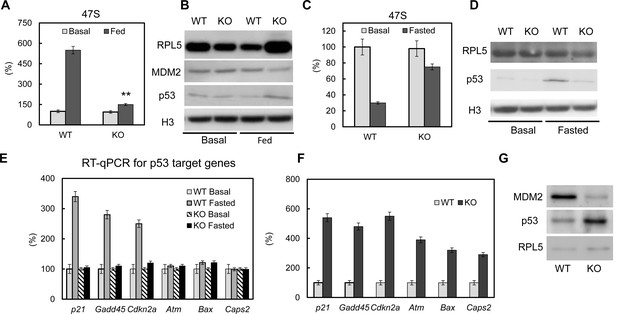
The p53 pathway is dys-regulated upon loss of PTRF.
Pre-rRNA levels from fed (A) and fasted (C) control and CRISPR/Cas9 edited PTRF null adipocytes were determined by RT-qPCR. Nucleus fraction lysates from fed (B) or fasted (D) adipocytes were separated in SDS-page followed by western blot by using the indicated antibodies, RPL5: ribosomal protein L5; MDM2: mouse double minute 2 homolog; H3: histone-H3. (E) p53 pathway related marker gene expression levels were measured from basal and fasted control or CRISPR/Cas9 edited PTRF null adipocytes by RT-qPCR. Gadd45: Growth arrest and DNA-damage-inducible protein GADD45; Cdkn2a: cyclin-dependent kinase inhibitor 2; Atm: Ataxia telangiectasia mutated; Bax: bcl-2-like protein 4; Caps2: Calcyphosine 2. (F) p53 pathway related marker gene expression levels were measured from six-month old wild type control and PTRF null mice adipose tissues. (G) Wild type and PTRF null mice adipose tissue lysate were subject to western blots by using indicated antibodies. **p<0.01; Student’s test. Error bars indicate SD.
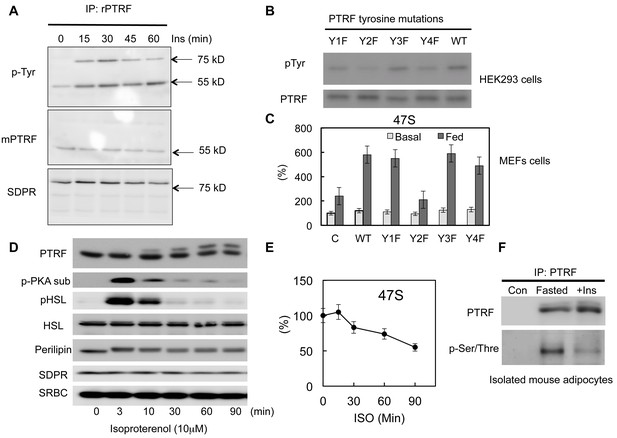
PTRF regulates ribosomal transcription through tyrosine and Ser/Thre phosphorylations.
(A) Isolated mouse primary adipocytes were stimulated by insulin from 0–60 min. The whole cell lysates were immunoprecipitated by PTRF antibody followed by SDS-PAGE and blotted with indicated antibodies. (B) Whole cell lysates from wild type PTRF and various single tyrosine mutants (1: Y13F; 2: Y158F; 3: Y310F; 4: Y318F) transfected HEK293 cell were immunoprecipitated by PTRF antibody followed by SDS-PAGE and blotted with indicated antibodies. (C) Basal and nutrients/insulin stimulated 47S levels were measured from control, wild type PTRF and various single tyrosine mutants transfected PTRF null MEFs cells. (D) Whole cell lysates from 3T3-L1 adipocyte stimulated by isoproterenol (ISO) for 0–90 min were subject to SDS-PAGA and blotted with indicated antibodies. (E) 47S levels were measured from ISO stimulated 3T3-L1 adipocytes by RT-qPCR. (F) Isolated mouse primary adipocyte were nutrients starved (fasted) or stimulated by insulin with nutrients. The whole cell lysates were immunoprecipitated by PTRF antibody followed by SDS-PAGE and blotted with indicated antibodies.
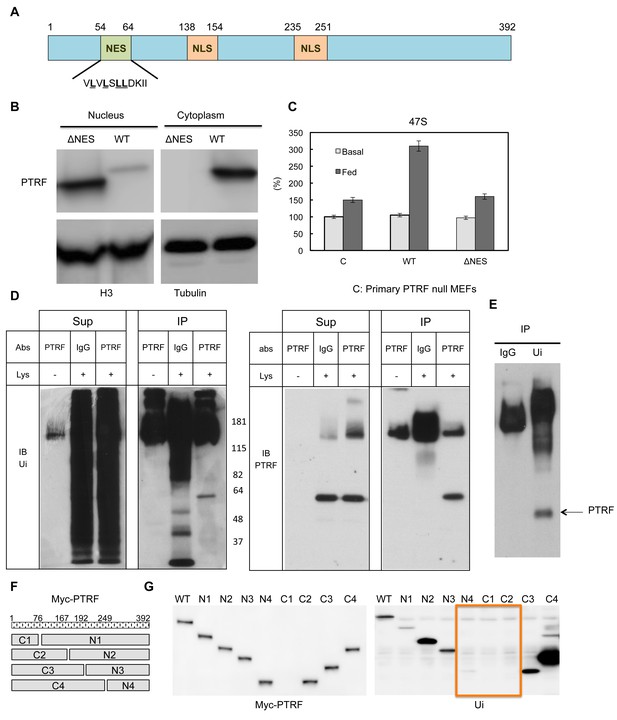
PTRF nuclear localization is regulated by its nuclear export signals and other post-translational modifications.
(A) Schematic illustration of nuclear export signals (NES) and nuclear localization signal (NLS) motifs in mouse PTRF protein. (B) NES deleted PTRF constructs and wild type control were transfected to HEK293 cells. The nuclear and cytoplasmic fractions were separated on SDS-PAGE following by immunoblot using indicated antibodies. (C) Basal and nutrients/insulin-stimulated 47S levels were measured from empty vector controls (C), wild type (WT) and NES motif deleted PTRF cDNA constructs transfected PTRF null MEFs cells. (D and E) Total rat adipocyte lysate were immunoprecipitated by IgG, or PTRF or ubiquitin (Ui) followed by SDS-PAGE and blotted with indicated antibodies. (F) A scheme of 8 designed PTRF fragment mutants (C1-4, N1-4). (G) Whole cell lysates from Wild type HEK293 cells transfected by (WT) and 8 PTRF fragment mutants were immunoprecipitated by myc-tag magnetic beads followed by immuno-blots with indicated antibodies.
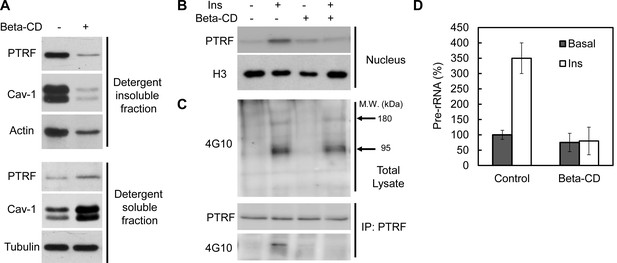
Intact caveolae are required for PTRF tyr-phosphorylation, nuclear translocation and ribosomal function.
(A) Isolated mouse primary adipocytes treated with beta-cyclodextrin (Beta-CD) or not were fractionated into crude lipid rafts and non lipid rafts fractions. Lysate from both fractions were run on SDS-PAGE and blotted with indicated antibodies. (B) Nuclear fraction lysates from beta-CD (45-min) and insulin (30-min) treated or not adipocyte were run on SDS-PAGE and blotted with indicated antibodies, histone-H3:H3. (C) Whole cell lysates from B were separated by SDS-PAGE and blotted with phosphotyrosine antibody (4G10). Arrows indicate the size of 180 and 95 kDa, which possibly corresponds to IRS-1/2 and beta-subunit of insulin receptor respectively. The whole cell lysates were also immunoprecipitated by PTRF antibody followed by SDS-PAGE and blotted with indicated antibodies. (D) 47S levels of the samples from B were measured by RT-qPCR.
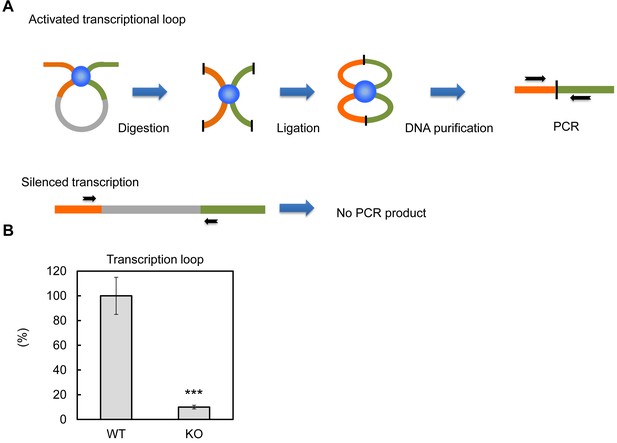
PTRF dependent active ribosomal transcription complex formation.
(A) Schematic illustration of chromatin confirmation capture (3C) assay, details are described in Method. (B) Following 3C assay, the relative levels of active transcription were quantified by RT-qPCR using nutrition/insulin stimulated primary mouse adipocytes from wild type (WT) and PTRF null (KO) mice (n = 4). ***p<0.001; Student’s test. Error bars indicate SD.
Additional files
-
Supplementary file 1
List of primers.
- https://doi.org/10.7554/eLife.17508.014





