Heterochromatin assembly by interrupted Sir3 bridges across neighboring nucleosomes
Figures
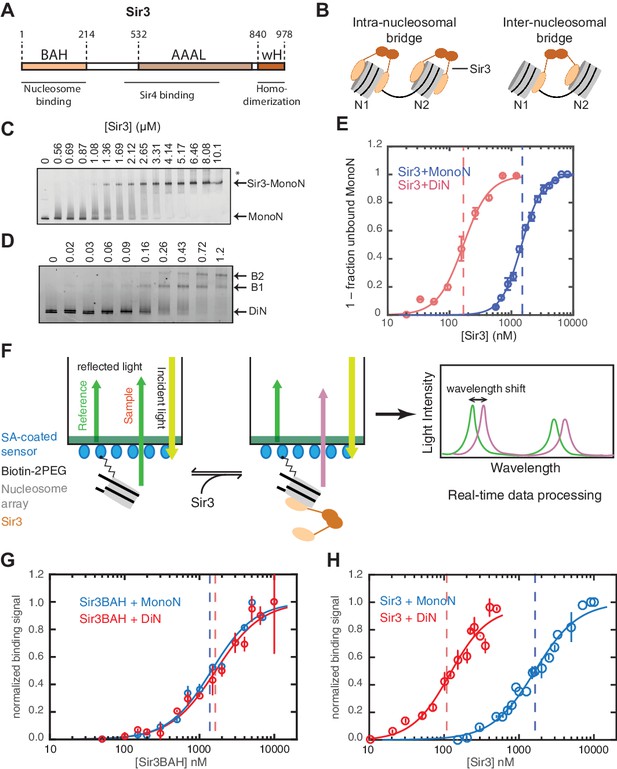
Cooperative association of Sir3 with DiN.
(A) Schematic diagram of the Sir3 primary sequence showing the location of the BAH, AAAL, and wH domains. (B) Models for the association of Sir3 dimers with chromatin. (C, D) Representative EMSA showing Sir3 binding to unmodified MonoN (C) and DiN (D). Purified Sir3 proteins were titrated onto a constant amount of MonoN or DiN at 3 nM. Samples were separated on native gels, nucleosomes were stained with SYBR Gold, and the amount of unbound nucleosomes was quantified by the staining intensity of the unshifted nucleosome band. *, higher mobility shifted band that may result from either bridging of MonoN by Sir3 or other minor high molecular weight Sir3-MonoN complexes. Band 1 (B1) likely reflects Sir3-DiN in bridged conformation, whereas Band 2 (B2) shows additional binding to single nucleosome surfaces. (E) Quantification and analysis of Sir3 binding to MonoN. Binding curves from three experiments performed as in (C) and (D) were fitted with the Hill Equation. The apparent KD values for Sir3 binding to MonoN (blue dotted line) and DiN (red dotted line) are indicated. (F) Bio-layer interferometry (BLI) assay detects changes in binding of proteins to surface-immobilized nucleosome templates by measuring the wavelength shift of the light reflected from the surface. (G, H) Binding of Sir3BAH (a.a. 1–382) (G) and full-length Sir3 (H) to MonoN and DiN after background correction and normalization to min-max of binding signal was fit with the Hill equation (see Materials and methods and Table 1). Data from 2–3 replicate experiments (>30 data points) (G) and from 3 or more replicate experiments (>30 data points) (H) were pooled for model fitting. The apparent KD for Sir3BAH and Sir3 binding to MonoN (blue) and DiN (red) is indicated. See Table 1 for parameter values.
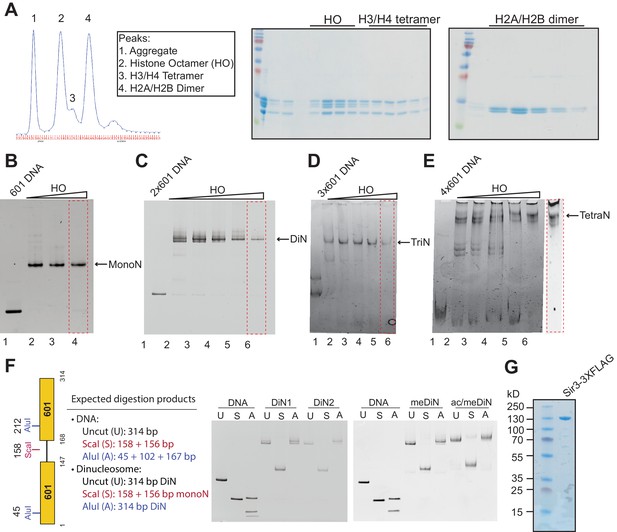
In vitro reconstitution of MonoN, DiN, TriN, and TetraN.
(A) Purification and analysis of the yeast histone octamer (HO) using gel filtration. HO refolded from individual histones was purified on a gel filtration column. Fractions from the gel filtration column (left) were run on denaturing SDS polyacrylamide gel (right). (B–E) Reconstitution of MonoN (B), DiN (C), TriN (D), and TetraN (E) using a titration of different HO:DNA ratios. The reconstitutions that produced a single sharp band (red dotted box) were used for biochemical assays. (F) Left: Illustration of the DiN DNA construct, and the predicted restriction enzyme digestion pattern for naked DNA and fully reconstituted DiN. Right: Restriction enzyme protection assay of reconstituted DiN. U: uncut, S: digested with ScaI, A: digested with AluI. Note that ScaI but not AluI digestion converts slower migrating DiN to faster migrating MonoN, whereas naked DNA is cleaved with both enzymes, as expected. (G) Purified Sir3-3XFLAG used in binding experiments.
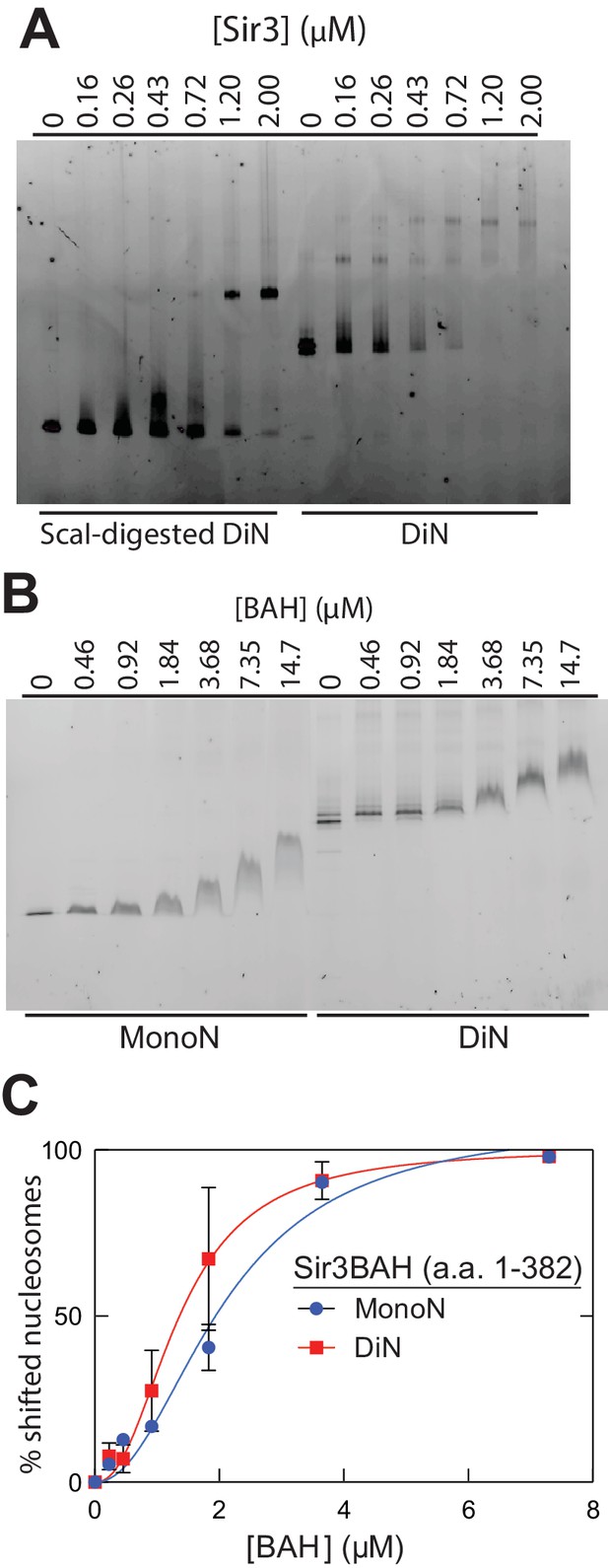
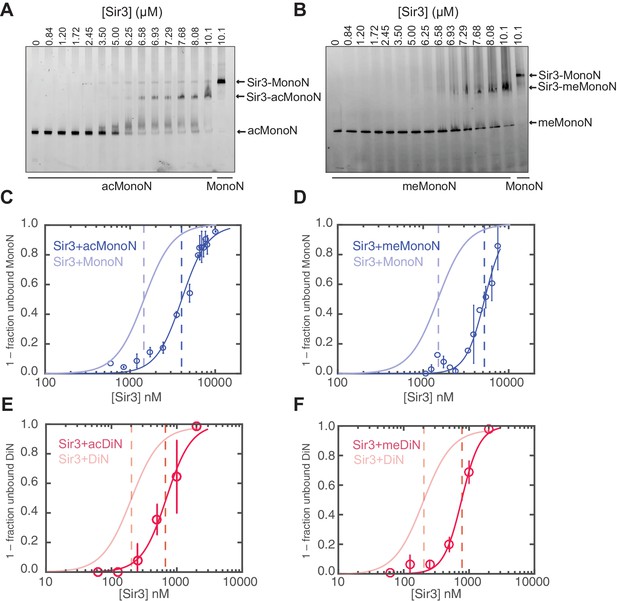
Association of the Sir3 with DiN and Sir3BAH domain with MonoN and DiN.
(A) EMSA experiments showing binding of the Sir3 domain to DiN with and without ScaI digestion. (B) EMSA experiments showing binding of the Sir3BAH domain to MonoN and DiN. The continuous up-shift of bands may be due to BAH-BAH interactions (Armache et al., 2011) or nonspecific BAH-MonoN interactions in gel buffer at high BAH concentrations. (C) Binding curves from three experiments were fitted with the Hill Equation.
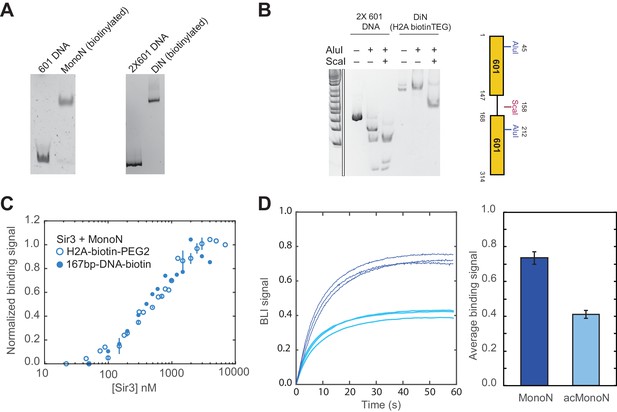
Measurements of Sir3 binding to MonoN with BLI.
(A) MonoN and DiN reconstitution using histone octamers containing histone H2A-K120C covalently linked to biotin-PEG2 maleimide. (B) Restriction enzyme protection assay of biotinylated DiN. (C) BLI assay of Sir3 binding to MonoN with differently biotinylated MonoN substrates yields identical results. The titration shows normalized equilibrium binding signal, integrating all modes of Sir3 binding to MonoN. H2A-biotin-PEG2 MonoN: Maleimide-PEG2-Biotin moiety covalently attached to histone H2A K120C SH group, 167bp-DNA-biotin: biotin moiety is covalently attached to the 5’-end at the beginning of DNA template (20 bp extension + 147 bp 601 positioning sequence). (D) Equilibrium binding signal to unmodified (dark blue) or acetylated (light blue) nucleosomes bearing H2A-biotin-PEG2 were measured by BLI at [Sir3] = 1.0–1.5 µM. Nucleosomes were loaded on streptavidin-coated BLI sensors to the same level before being used for Sir3 titration experiments. Histograms show the average and standard deviation of the equilibrium binding signal of the three BLI sensograms shown on the left.
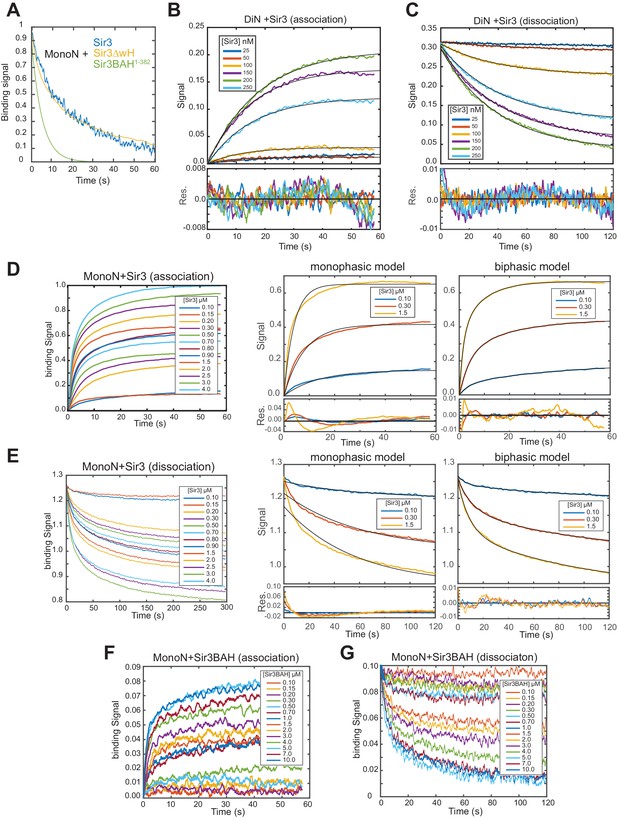
Analysis of BLI binding profiles.
(A) Representative Sir3-MonoN complex decay curves for full-length (Sir3), Sir3∆wH, and BAH (Sir3 a.a. 1–382) proteins indicate that deletion of AAA-ATPase like domain, unlike the wH domain has a strong effect on nucleosome residence time of Sir3. For Sir31–382, best fit to dissociation curve is shown instead of raw data. Dissociation curves are normalized to their ranges for easier visual comparison. (B, C) Real-time association (B) and dissociation (C) profiles of Sir3 binding to unmodified DiN at indicated concentrations of Sir3. Higher concentrations are not shown to highlight the range of concentrations in which Sir3 binds cooperatively to di-nucleosome. The black lines indicate the best fit mono-exponential models (see Materials and methods) with associated fit residuals shown in the bottom plot. (D, E) Real-time association (D) and dissociation (E) profiles of Sir3 binding to unmodified MonoN at indicated concentration of Sir3. Fits with mono- and bi-exponential models (black lines, see Materials and methods) at representative Sir3 concentrations are shown on the right. The systematic deviation of the mono-exponential model from data results in a periodic (non-random) change of sign in residuals (bottom plots) between positive and negative values, indicating model inadequacy. Fits with bi-exponential model show a substantial decrease and random distribution around zero of fit residuals. (F, G) Real-time association (F) and dissociation (G) profiles of Sir3BAH binding to unmodified MonoN at indicated concentrations of Sir3BAH showing mono-phasic association and dissociation. In protein concentrations above 1 µM a slow association/dissociation phase with small amplitude appeared, which likely reflected nonspecific interactions with nucleosomes. Measurements at different protein concentrations in B–G are performed by separate BLI sensors, pre-loaded with biotinylated nucleosomes to identical levels in each titration (± 5% variation). Dissociation profiles (C, E, G) are aligned at time zero to an arbitrary value.
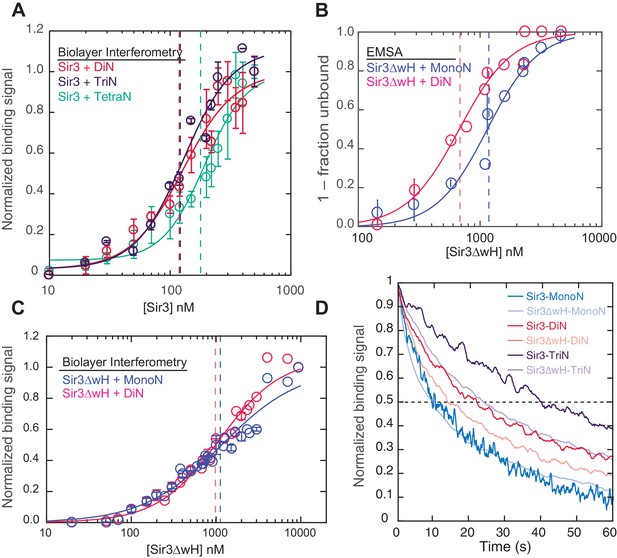
Sir3 displays maximal cooperative binding to DiN that is mediated by its winged helix (wH).
(A) Binding of full-length Sir3 to DiN, TriN, and TetraN templates after background correction and normalization to min-max of binding signal was fit with the Hill equation (see Materials and methods and Table 1). Data from 3 or more replicate experiments (>30 data points) were pooled for model fitting. Vertical dotted line indicates the apparent KD for Sir3 binding to DiN, TriN and TetraN. (B) EMSA showing the binding of Sir3ΔwH to MonoN (blue) and DiN (red). Binding curves from three experiments performed (see Figure 2—figure supplement 1) were fitted with the Hill Equation. (C) Sir3ΔwH MonoN and DiN binding data from BLI assays normalized to the range of binding signal and fit with Hill equation (see Materials and methods). Data from two independent replicates were pooled before model fitting. (D) Kinetics of Sir3-nucleosome complex dissociation measured by the BLI assay reveals that the Sir3wH domain is required for the cooperative stabilization of Sir3-DiN and Sir3-triN complexes. Representative dissociation profiles obtained at 0.2 µM Sir3 were self-normalized for easier visual comparison. See Table 1 for parameter values.
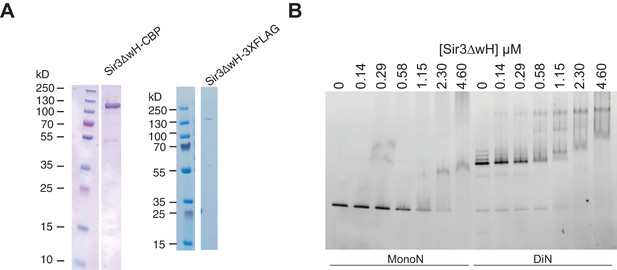
Purification of Sir3∆wH and EMSA assays with Sir3∆wH.
(A) Coomassie-stained gels showing purified Sir3ΔwH-CBP (left) and Sir3ΔwH-3XFLAG (right) proteins used in EMSA and BLI experiments. (B) EMSA experiments comparing the binding of Sir3ΔwH-3XFLAG to unmodified MonoN or DiN.
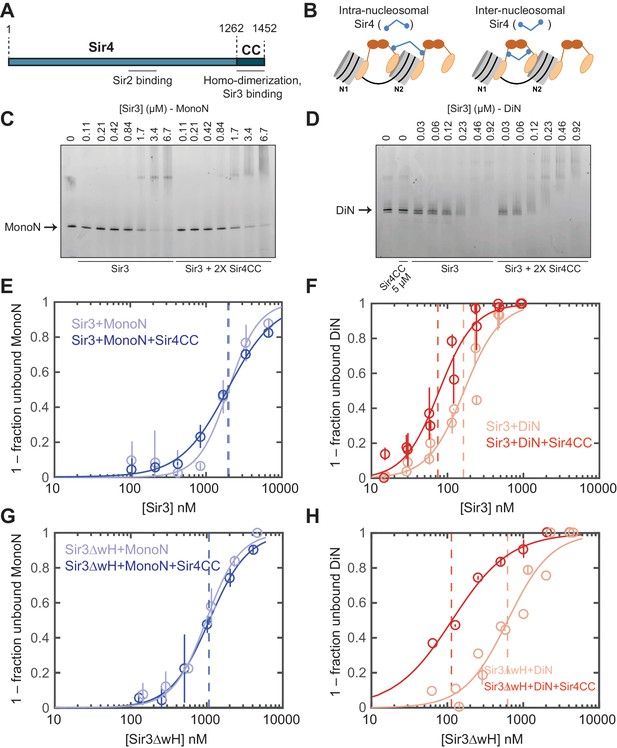
Sir4CC does not affect Sir3 binding to MonoN, but increases its affinity towards DiN.
(A) Schematic diagram of Sir4 primary sequence showing the location of the coiled-coil (CC) domain and the Sir2 interaction domain (aa747–893). (B) Models for the association of Sir4 with Sir3-bound nucleosomes. (C, D) EMSA experiments comparing Sir3 binding to MonoN (C) and DiN (D) in the presence or absence of Sir4CC. (E, F) Binding curves from three experiments performed as in (C) and (D), respectively, were fitted with the Hill Equation. (G, H) Comparison of Sir3ΔwH binding to MonoN (G) and DiN (H) in the presence or absence of Sir4CC. Binding curves from three experiments performed as in Figure 3—figure supplement 1A and B were fitted with the Hill equation.
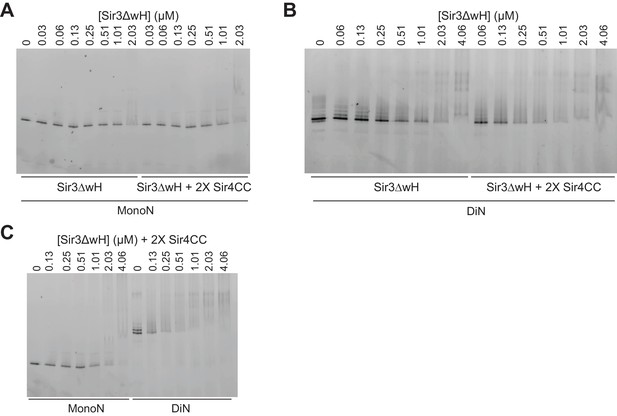
EMSA assays with Sir3∆wH and Sir4CC.
(A) EMSA experiments comparing the binding of Sir3ΔwH to unmodified MonoN in the presence or absence of 2X molar excess of Sir4CC. Quantitative analysis of the results is presented in Figure 3G. (B) Comparison of Sir3ΔwH binding to DiN in the presence or absence of 2X molar excess of Sir4CC. Quantitative analysis of the results is presented in Figure 3H. (C) Comparison of Sir3ΔwH binding to MonoN and DiN in the presence of 2-fold molar excess of Sir4CC.
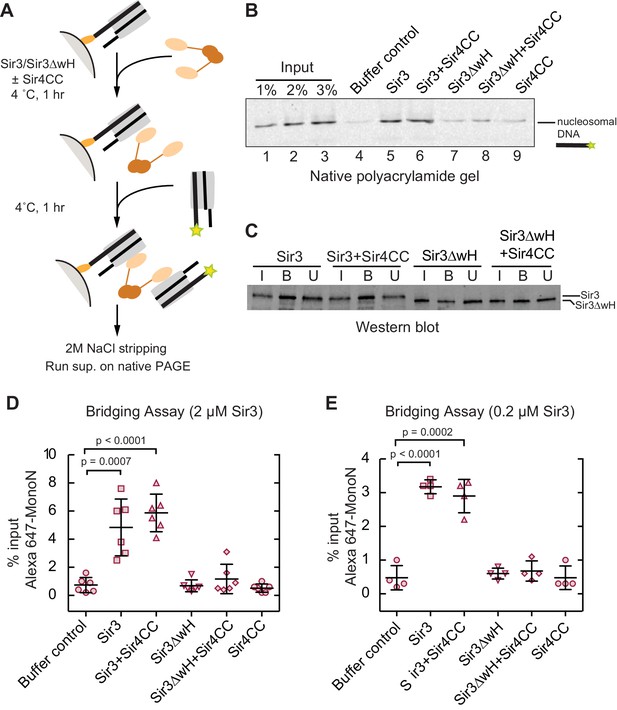
Sir3 crosslinks free mono-nucleosomes in solution.
(A) Illustration of the crosslinking assay. (B) A representative native polyacrylamide gel showing the fluorescent DNA pulled down from different reaction mixtures. (C) A representative western blot showing Sir3 and Sir3ΔwH in all reactions bound to nucleosomes efficiently. I, input; B, bound, U, unbound. (D) Percentage of input Alexa 647- nucleosomes that was pulled down in different reaction mixtures where 2 µM Sir3 or Sir3ΔwH protein concentration was used. Quantification for 6 experiments is presented. (E) Same as D but using 0.2 µM Sir3 or Sir3ΔwH protein. Quantification for 4 experiments is presented.
H4K16 acetylation and H3K79 methylation act together to strongly inhibit the binding of Sir3 to nucleosomes.
(A, B) Comparison of Sir3 binding to unmodified and H4K16ac MonoN (A), or unmodified and H3KC79me3 MonoN (B) by EMSA. We note that Sir3 bound H4K16ac and H3KC79me3 MonoN and DiN shift to a lower position than Sir3 bound unmodified nucleosomes, suggesting that Sir3 binds to modified nucleosomes in a different conformation than to unmodified nucleosomes. (C) Binding curves from three experiments performed as in (A) were fitted with the Hill Equation. (D) Binding curves from three experiments performed as in (B) were fitted with the Hill Equation. (E) Comparison of Sir3 binding to unmodified and H4K16ac DiN. Binding curves from three experiments performed as in Figure 5—figure supplement 2A were fitted with the Hill Equation. (F) Comparison of Sir3 binding to unmodified and H3KC79me3 DiN. Binding curves from three experiments performed as in Figure 5—figure supplement 2B were fitted with the Hill Equation. In C–F, curves in lighter colors show model fits to binding data of unmodified nucleosomes for visual comparison. Data and fits are shown in Figure 1E. Blue and Red dotted lines indicate the apparent KD for Sir3 binding to MonoN and DiN, respectively. See Table 1 for parameter values. Note that the titrations of modified nucleosomes are not fully saturated at the highest concentrations tested and the calculated apparent affinities may be overestimated.
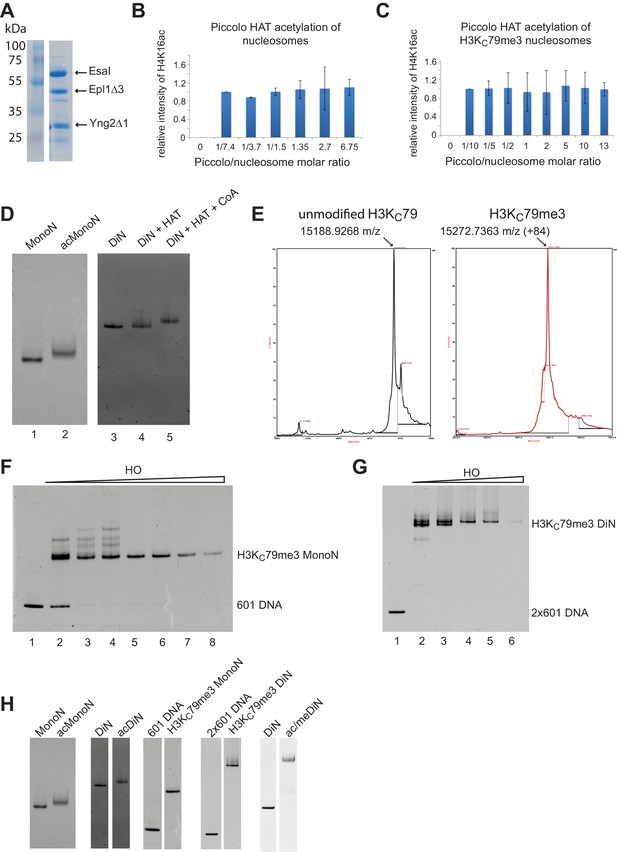
Reconstitution of H4K16ac and H3KC79me3 nucleosomes.
(A) Coomassie stained gel showing purified Piccolo HAT complex. (B) Quantification, by quantitative Western Blot, of Piccolo HAT mediated acetylation on unmodified nucleosomes. Increased amounts of Piccolo HAT complex were titrated against a constant amount of unmodified nucleosomes. The resulting H4K16ac level was quantified using anti-H4K16ac antibody. There was no increase in acetylation signal with increasing amount of Piccolo HAT, indicating saturating levels of acetylation. (C) Quantification of acetylation reactions as in (B) but on H3KC79me3 nucleosomes. (D) Nucleosome acetylation causes a slight shift of nucleosome, consistent with previous findings. Modified nucleosomes are completely shifted, indicating complete acetylation. (E) Mass Spectrometry verified the complete alkylation of H3KC79me3. (F) Reconstitution of H3KC79me3 MonoN. (G) Reconstitution of H3KC79me3 DiN. Nucleosome reconstitutions that produced a single band were used for biochemical assays. (H) Assembly of unmodified, singly, or doubly modified mono- and DiN. H4K16ac DiN gel was taken from lanes 5 and 7 in (D); H3KC79me3 MonoN gel was taken from lanes 1 and 6 in (F); H3KC79me3 DiN gel was taken from lanes 1 and 5 in (G).
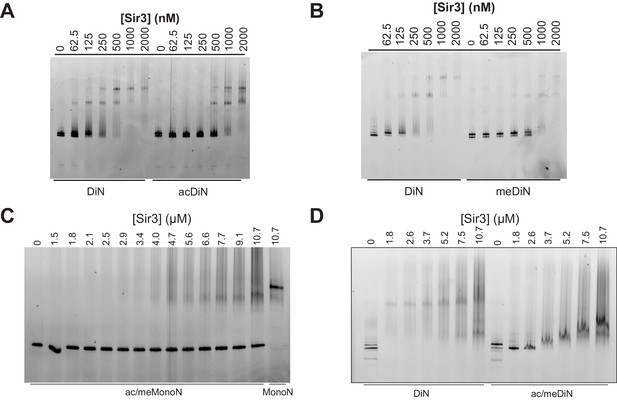
Sir3 binding to singly and doubly modified H4K16 acetylated and H3K79 methylated MonoN and DiN.
(A) EMSA experiments comparing Sir3 binding to the unmodified and H4K16ac DiN. (B) EMSA experiments comparing Sir3 binding to unmodified and H3KC79me3 DiN. We note that Sir3 bound H4K16ac and H3KC79me3 MonoN and DiN (Figure 5A–B) shift to a lower position than Sir3 bound unmodified nucleosomes, suggesting that Sir3 binds to modified nucleosomes in a different conformation than to unmodified nucleosomes. (C) EMSA experiments showing Sir3 binding to doubly modified H4K16ac/H3KC79me3 MonoN. There was about 10% MonoN shift at 10.7 μM Sir3. (D) EMSA experiments showing Sir3 binding to unmodified and doubly modified H4K16ac/H3KC79me3 DiN.
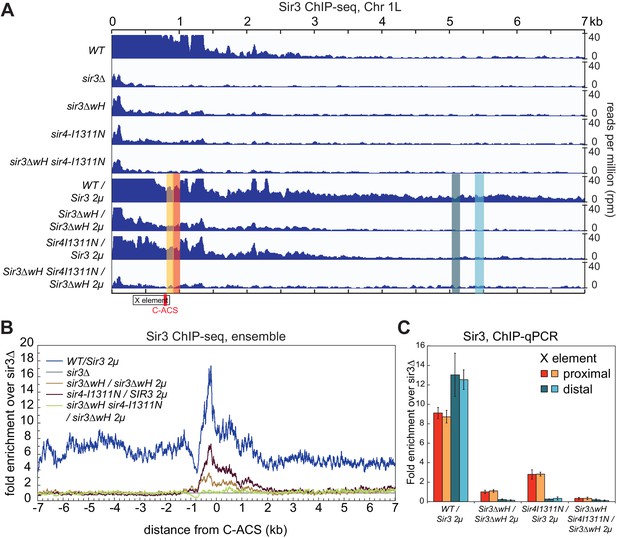
Sir3wH and Sir4CC are both required for SIR complex spreading in vivo but play partially redundant roles when Sir3 is overexpressed.
(A) Normalized ChIP-seq read densities for Sir3 at the left telomere of chromosome 1 (Chr1L) from cells with the indicated genotypes. The core X element (black box) and the ACS within (C-ACS filled red box) are indicated below the tracks. Sir3 was overexpressed from a 2 micron plasmid (SIR3 2 µ). Colorshaded areas indicate regions assayed by ChIP-qPCR, shown in panel C. Full scale plots of WT and WT/Sir3 2µ strains are shown in Figure 6—figure supplement 1B. (B) Ensemble plot of Sir3 ChIP-seq signals with Sir3 overexpression, from 30 subtelomeres, excluding TEL01R and TEL13R, aligned at C-ACS. ChIP signal at all 30 subtelomeric regions was summed up, normalized to the sir3Δ sample, and plotted as a function of distance from C-ACS. Negative values are towards the chromosome end, and positive values are towards the centromere. Similar plot with endogenous Sir3 expression level is shown in Figure 6—figure supplement 1A. (C) Sir3 ChIP-qPCR results from cells with the indicated genotypes normalized to sir3∆ cells. Error bars indicate the standard deviation of three biological replicates. PCR primer sets (Table 4) were chosen to assay regions shown with corresponding colors in panel A.
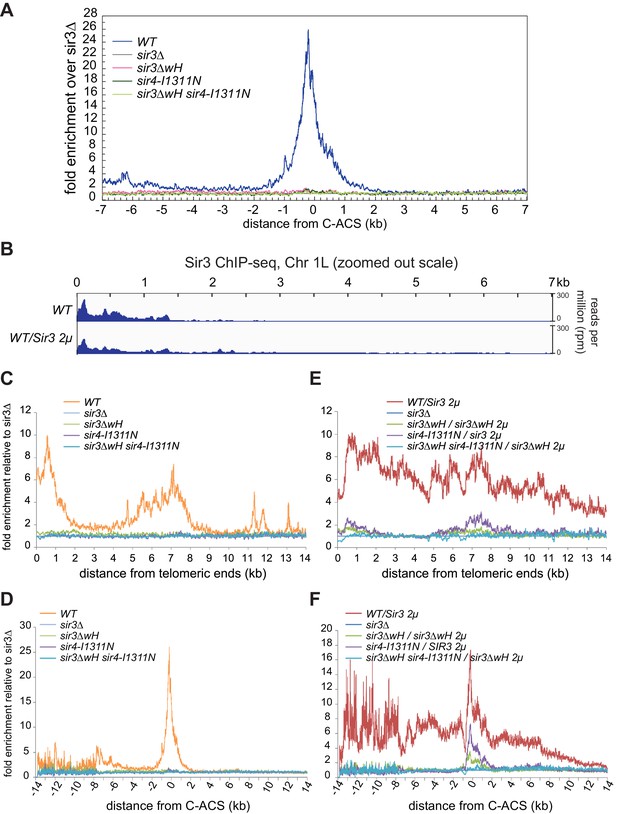
The requirement of Sir3wH and Sir4CC for the association of Sir3 with heterochromatin in vivo.
(A) Ensemble plot of Sir3 ChIP-seq signals with endogenous Sir3 expression level, from 30 subtelomeres, excluding TEL01R and TEL13R, aligned at C-ACS. Negative values are towards chromosome end, and positive values are towards centromere. (B) Normalized ChIP-seq read densities for Sir3 and overexpressed Sir3 (Sir3 2 µ) at the left telomere of chromosome 1 (Chr 1 L). Full-scale plot for comparison with Figure 6A. (C, E) Ensemble plots of Sir3 ChIP-seq signals, without (C) and with (E) Sir3 overexpression, from 30 subtelomeres, excluding TEL01R and TEL13R, aligned at chromosome ends. ChIP signal at all 30 subtelomeric regions was summed up, normalized to the sir3Δ sample, and plotted as a function of distance from chromosome end. (D, F) Ensemble plots of Sir3 ChIP-seq signals, without (D) and with (F) Sir3 overexpression, from 30 subtelomeres, excluding TEL01R and TEL13R, aligned at C-ACS. Negative values are towards the chromosome end, and positive values are towards the centromere. ChIP signal at all 30 subtelomeric regions was summed up, normalized to the signal from sir3Δ cells, and plotted as a function of distance from C-ACS.
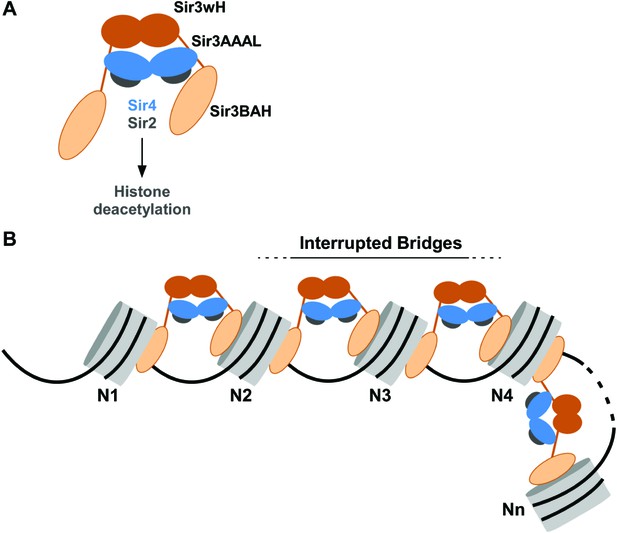
Association of the SIR complex with chromatin by interrupted Sir3 bridges across neighboring nucleosomes.
(A) Schematic of the SIR complex indicating the location of domains in Sir3 (brown) and Sir4 (blue) dimers, and highlighting the interaction of Sir4 with Sir2. Not to scale. (B) Model for the association of the SIR complex with chromatin. Sir3 dimers form inter-nucleosomal bridges via cooperative association with histone H4K16 and H3K79 regions on flat nucleosome surfaces. The Sir3 bridge requires the wH domain and is further stabilized via interactions between the coiled-coil domain of Sir4 and the AAAL domain of Sir3. Spreading does not involve interaction of newly recruited Sir3/Sir4 with already bound complexes but instead requires cooperative association with a pair of H4K16 deacetylated and H3K79 unmethylated nucleosome surfaces, and thus is driven primarily by Sir2-dependent deacetylation of proximal nucleosomes. The inability of Sir3 to simultaneously interact with binding sites on the same nucleosome, requires spreading via the formation of interrupted Sir3 bridges. Sir3 may interact with immediately neighboring nucleosomes (N1 and N2, N2 and N3, N3 and N4) or more distal nucleosome pairs lacking H4K16ac/H3K79me modifications (N4 and Nn).
Tables
Thermodynamic parameters describing the binding of Sir3 protein to nucleosomes. Data from more than 2 replicate titration experiments were pooled and the BLI data were fit with Hill equation (see Materials and methods). Uncertainties show 68% confidence intervals around fit parameters (±1 SD) reported by fitting algorithm.
Binding experiments | BLI | EMSA* | ||
|---|---|---|---|---|
Apparent KD (µM) | Hill coefficient | Apparent KD (µM) | ||
MonoN | Sir3 | 1.4 ± 0.06 | 1.3 ± 0.1 | 1.7 ± 0.20 |
Sir3+Sir4CC | N/A | N/A | 1.4 ± 0.10 | |
Sir3∆wH | 1.2 ± 0.10 | 0.93 ± 0.07 | 1.0 ± 0.10 | |
Sir3∆wH+Sir4CC | N/A | N/A | 0.9 ± 0.05 | |
Sir3BAH | 1.4 ± 0.10 | 1.5 ± 0.2 | 2.1 ± 0.20 | |
DiN | Sir3 | 0.12 ± 0.01 | 1.9 ± 0.2 | 0.17 ± 0.10 |
Sir3+Sir4CC | N/A | N/A | 0.08 ± 0.01 | |
Sir3∆wH | 1.1 ± 0.05 | 1.2 ± 0.1 | 0.62 ± 0.10 | |
Sir3∆wH+Sir4CC | N/A | N/A | 0.12 ± 0.01 | |
Sir3BAH | 1.6 ± 0.10 | 1.4 ± 0.1 | 1.40 ± 0.20 | |
Sir3 | acMonoN | N/A | N/A | 4.0 ± 0.20 |
meMonoN | N/A | N/A | 5.2 ± 0.20 | |
acDiN | N/A | N/A | 0.7 ± 0.05 | |
meDiN | N/A | N/A | 0.8 ± 0.05 | |
ac/meMonoN | N/A | N/A | >11† | |
ac/meDiN | N/A | N/A | >3† | |
-
*Hill coefficients obtained from EMSA appeared unreliable due to assay artifacts, such as non-specific binding to DNA, and are not reported.
-
† Nonspecific binding could not be measured accurately due to low affinity.
Kinetic parameters describing the binding of Sir3 protein to nucleosomes. Rates and amplitudes, representing the average of 3 or more measurements at different Sir3 concentrations in the ranges specified below, were obtained by fitting data with appropriate rate equations (Suppl. Materials and methods). Values in parentheses indicate standard deviations. In the low range of concentrations, binding rates to mono- and di-nucleosomes were within the expected range of diffusion-limited rate of protein interactions (Schreiber et al., 2009). At higher concentrations, however, binding proceeded with rates slower than diffusion limit, probably due to the competition with other modes of binding to nucleosome surfaces.
Association | Aobs, 1 | kobs, 1 (s−1) | Aobs, 2 | kobs, 2 (s−1) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
[Sir3] < 0.3 µM | MonoN | Sir3 | 51% | (11%) | 0.25 | (0.07) | 49% | (11%) | 0.04 | (0.01) |
Sir3∆wH | 73% | (11%) | 0.14 | (0.06) | 27% | (11%) | 0.03 | (0.01) | ||
Sir3BAH | 100% | (0%) | 0.65 | (0.34) | 0% | (0%) | N/A | N/A | ||
DiN | Sir3 | 100% | (0%) | 0.06 | (0.01) | 0% | (0%) | N/A | N/A | |
Sir3∆wH | 11% | (22%) | 0.16 | N/A | 89% | (22%) | 0.08 | (0.01) | ||
Sir3BAH | 100% | (0%) | 0.40 | (0.18) | 0% | (0%) | N/A | N/A | ||
[Sir3] 1.5–4 µM | MonoN | Sir3 | 56% | (1%) | 0.62 | (0.02) | 44% | (1%) | 0.08 | (0.01) |
Sir3∆wH | 74% | (4%) | 0.70 | (0.07) | 26% | (4%) | 0.10 | (0.02) | ||
Sir3BAH* | 100% | (0%) | 0.53 | (0.06) | 0% | (0%) | N/A | N/A | ||
DiN | Sir3 | 55% | (2%) | 0.51 | (0.10) | 45% | (2%) | 0.08 | (0.00) | |
Sir3∆wH | 56% | (3%) | 0.47 | (0.05) | 44% | (3%) | 0.09 | (0.02) | ||
Sir3BAH* | 100% | (0%) | 0.46 | (0.03) | 0% | (0%) | N/A | N/A | ||
i † | Aoff, 1 | τoff, 1(S) | Aoff, 2 | τoff, 2 (S) | kon,1 (M−1s−1)‡ | kon,2 (M-1s−1)‡ | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
[Sir3] < 0.3 µM | MonoN | Sir3 | 2 | 38% (3%) | 7.4 (3.5) | 62% (3%) | 78.5 (23.7) | 2.4E + 05 | (6.5E + 4) | 6.9E + 04 | (2.0E + 4) |
Sir3∆wH | 2 | 56% (2%) | 8.4 (0.9) | 44% (2%) | 75.3 (3.6) | 1.1E + 0 | (4.3E + 4) | 3.6E + 04 | (1.4E + 4) | ||
Sir3BAH | 2 | 100% (0%) | 5.6 (1.9) | 0% (0%) | N/A N/A | 1.4E + 06 | (9.2E + 5) | N/A | N/A | ||
DiN | Sir3 | 1 | 92% (14%) | 51.6 (3.8) | 8% (14%) | N/A N/A | 3.8E + 05 | (6.8E + 4) | N/A | N/A | |
Sir3∆wH | 4 | 52% (2%) | 10.8 (1.2) | 48% (2%) | 83.8 (6.5) | 9.7E + 04 | (3.7E + 4) | 4.4E + 04 | N/A | ||
Sir3BAH | 4 | 100% (0%) | 5.0 (1.4) | 0% (0%) | N/A N/A | N/A | N/A | N/A | N/A | ||
[Sir3] 1.5–4 µM | MonoN | Sir3 | 2 | 52% (3%) | 7.7 (0.5) | 48% (3%) | 82.2 (3.7) | 9.1E + 04 | (2.4E + 4) | 1.3E + 04 | (4.0E + 3) |
Sir3∆wH | 2 | 69% (2%) | 3.4 (0.5) | 31% (2%) | 44.0 (12.4) | 8.5E + 04 | (3.2E + 4) | 1.6E + 04 | (7.3E + 3) | ||
Sir3BAH | 2 | 100% (0%) | 5.2 (0.5) | 0% (0%) | N/A N/A | 5.5E + 04 | (1.3E + 4) | N/A | N/A | ||
DiN | Sir3 | 4 | 42% (3%) | 5.8 (0.1) | 58% (3%) | 48.6 (2.8) | 3.7E + 04 | (8.1E + 3) | 7.2E + 03 | (3.5E + 3) | |
Sir3∆wH | 4 | 63% (1%) | 5.2 (0.6) | 37% (1%) | 48.5 (3.8) | 3.2E + 04 | (7.5E + 3) | 8.7E + 03 | (2.9E + 3) | ||
Sir3BAH | 4 | 100% (0%) | 5.2 (0.5) | 0% (0%) | N/A N/A | 2.1E + 04 | (7.1E + 3) | N/A | N/A | ||
-
* Slow dissociation phase was not quantified due to small amplitude.
-
† Presumed number of binding sites used in the calculation of kon.
-
‡ Rates are calculated from (kobs,1, koff,1) and (kobs,2, koff,2) pairs.
List of yeast strains and plasmids used in this study.
Name | Yeast strain genotype | Source |
|---|---|---|
W303-1a (SF1) | MATa ade2-1 can1-100 his3-11 leu2-3,112 trp1 ura3-1 GAL | J. Rine |
SF4 | sir3△::TRP1 in SF1 | J. Rine |
DMY4350 | sir3△wH::TRP1 in SF1 | This Study |
ADR2973 | sir4-I1311N in SF1 | |
DMY4351 | sir4-I1311N sir3△wH::TRP1 in SF1 | This Study |
DMY3315 | W303-1a sir3Δ::KanR hmrΔE::TRP1 TELVII-L::URA3 |
Name | Plasmid genotype | Source |
|---|---|---|
pRS315 | CEN/ARS LEU2 | |
pJR104 (pDM602) | SIR3 under endogenous promoter in YEp24 | |
pDM1798 | sir3△wH under endogenous promoter in YEp24 | This Study |
pDM832 | Sir3-3XFLAG under endogenous promoter in pRS315 | |
pDM1799 | Sir3△wH-3XFLAG under endogenous promoter in pRS315 | This Study |
-
All deletions and mutations were confirmed by PCR and sequencing. Epitope-tagged strains were constructed by a PCR-based gene targeting method (Longtine et al., 1998; Rudner et al., 2005).
List of qChIP PCR primers used in this study.
Primer | Sequence |
|---|---|
RB139 (C-ACS distal 1, F) | TTC TGC CCA TAC GAT ACC T |
RB140 (C-ACS distal 1, R) | AGT TAC GCG TGC TAC ATT AC |
RB141 (C-ACS distal 2, F) | GTT CTA CTG ACA GGA TGG AAT AG |
RB142 (C-ACS distal 2, R) | GTG AAG GAG GGC ATG AAA T |
RB143 (C-ACS proximal 1, F) | CGT ACT TAC ACA GGC CAT AC |
RB144 (C-ACS proximal 1, R) | GTT TGA GCC ACT ACC GTA TTA |
RB145 (C-ACS proximal 2, F) | CTT GTG GTA GCA ACA CTA TCA |
RB146 (C-ACS proximal 2, R) | GGC CTG TGT AAG TAC GAA AT |





