A cellular and regulatory map of the GABAergic nervous system of C. elegans
Figures

Anti-GABA staining defines the GABAergic nervous system of C. elegans.
(A) Three different optical sections of the head of a single adult hermaphrodite, stained with anti-GABA antibodies. Previously identified GABA neurons (McIntire et al., 1993b) are shown in bold. (B) Anti-GABA staining in the tail of an adult hermaphrodite. (C) Anti-GABA staining in the head of an adult unc-25/GAD(-) hermaphrodite. Absence of GABA staining illustrates that GABA staining is specific. Staining in muscles is diminished but not eliminated completely. (D) Two different optical sections (ventral/dorsal) and a lateral view of the head of a GABA-stained adult unc-47/VGAT(-) hermaphrodite, which illustrates the dependence of some GABA-positive cells on vesicular GABA secretion. Only the newly identified GABA neurons are labeled. SMD is more strongly stained in unc-47 mutants compared to wild type animals. The hmc, ALA and AVF are not stained. We also observed weak GABA staining in the IL1 neurons of unc-47(-) animals, but not in wild-type animals. (E) Two different optical sections of the head of a GABA-stained adult snf-11/GAT(-) hermaphrodite, which illustrates the dependence of some GABA-positive cells on GABA uptake. Only the newly identified GABA neurons are labeled. hmc, GLR, ALA and AVF are not stained. (F) Anti-GABA staining of AVF neurons is absent in unc-30 mutant animals. (A–F) (Scale bar = 10 μm)
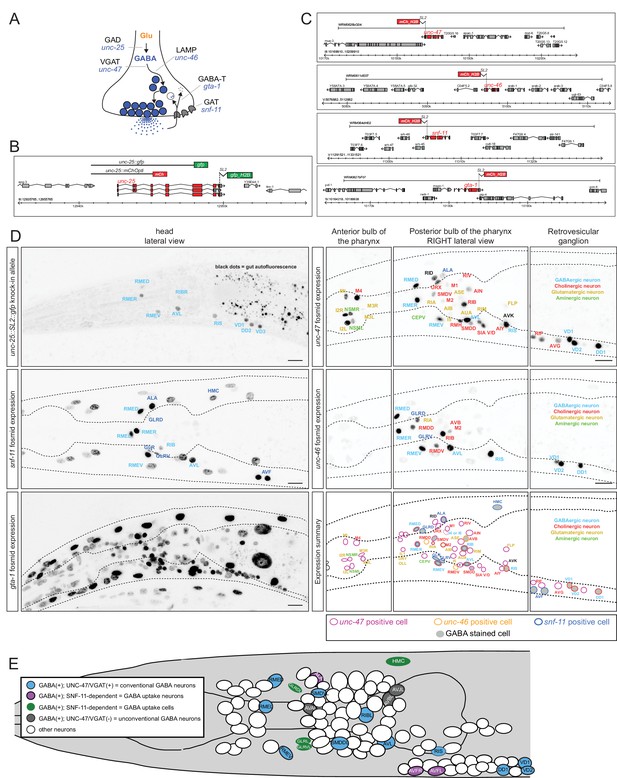
Reporter gene constructs recapitulate GABA antibody staining in the nervous system of the C. elegans hermaphrodite.
(A) GABA synthesis, secretion, uptake and degradation pathway. (B) unc-25 reporter transgene schematics and the unc-25 locus with the knock-in allele ot867[unc-25::SL2::gfp] which is targeted to the nucleus to facilitate the detection and the identification of expressing cells. (C) Fosmid-based reporter transgenes for unc-46, unc-47, snf-11 and gta-1. Note that the reporters are targeted to the nucleus to facilitate the identification of expressing cells. (D) Reporter expression. Images are color-inverted. The last panel shows a summary of all reporter expressing neurons. Font color indicates the known neurotransmitter used by the neuron, the colored circle which gene is expressed in the neuron and the grey shade if the neuron is GABA stained. (Scale bar = 10 μm) (E) Summary of all GABA-positive cells in the head of the worm (left view). Color coding same as in Table 1. Pharyngeal neurons are not shown. In the remainder of the body of the hermaphrodite, only the D-type motor neurons (some shown here) and the DVB tail neuron are GABA-positive. Dashed circle indicates neurons that present only on the right side of the animals, but shown here on the left side.

GABAergic neurons in the male.
(A–C) Anti-GABA staining of adult males in the head and tail in different optical sections. (Scale bar = 10 μm) (A) In adult male head, same neurons as in the hermaphrodite head are GABA-positive. (B) Seven male-specific neurons are GABA-stained: EF1-4, R2A, R6A and R9B (white arrows; two optical section with green fluorescent secondary antibody in left two panels with ray neuron staining being background and red fluorescent secondary antibody in upper right panels with ray neuron showing no background staining). (C) In unc-47 mutants, GABA staining in R2A, R6A and R9B persists, indicating that these neurons do not acquire GABA via secretion from other neurons. In addition, we observe GABA staining in an extra ray neuron, R3, that we cannot detect in wild-type animals. (D) Reporter gene expression in adult male tail. Left panel: unc-25 reporter, other panel shows red fosmid reporter (nucleus) and green anti-GABA staining. Reporter constructs as shown in Figure 2. EF1-4 expressed unc-25/GAD, unc-47/VGAT and snf-11/GAT. unc-47/VGAT is expressed in R6A but not in R2A or R9B. White arrow indicates GABA stained cells colocalizing with the reporter. snf-11/GAT is present in the male VD12 but not in the hermaphrodite. unc-25/GAD transcriptional reporter shows the same expression pattern as the gfp knock-in allele in the male tail (Scale bar = 10 μm).
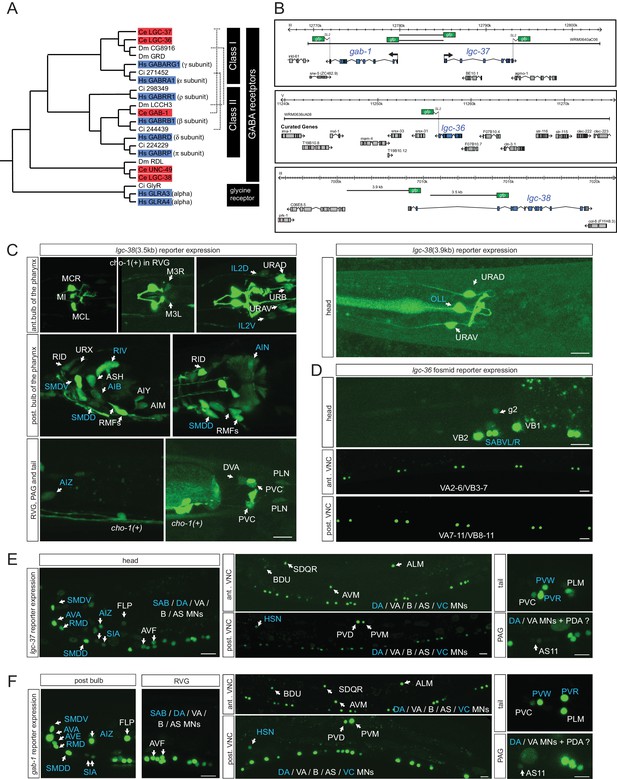
Ionotropic GABA receptor expression.
(A) Phylogeny of the C. elegans GABAA receptor-encoding genes (red boxes) that are most similar to canonical human GABAA receptor-encoding genes (blue boxes). The dendrogram has been adapted from (Tsang et al., 2007). Two other bona-fide GABA receptors, EXP-1 and LGC-35, are more distantly related, falling outside this part of the tree. (B) GABA reporter transgene schematics. All the fosmids are targeted to the nucleus to facilitate the detection and the identification of expressing cells. (C–F) Expression of lgc-38 (C), lgc-36 (D), lgc-37 (E) and gab-1 (F) reporter constructs. Blue labels indicate neurons innervated by a GABA-positive neuron (see Table 3). Cells were identified with unc-47, eat-4 or cho-1 reporters as landmarks in the background. The transcriptional reporter gene fusions for lgc-38 (see Materials and methods) do not capture the entire locus and hence they may lack regulatory information but there is no fosmid available that encompasses the entire locus. (Scale bar = 10 μm, VNC: ventral nerve cord, PAG: pre-anal ganglion, RVG: retro-vesicular ganglion; MN = motor neuron)
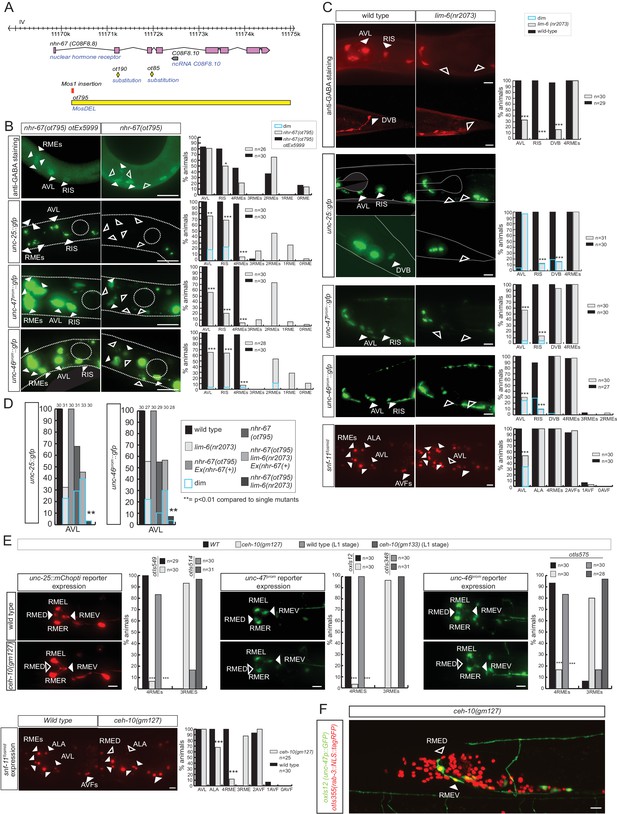
nhr-67 cooperates with distinct homeodomain proteins in distinct GABAergic neuron types.
(A) nhr-67 locus with newly generated null allele. (B–C) Anti-GABA staining and expression of GABA pathway genes in nhr-67 and lim-6 null mutant backgrounds (Fisher exact test for all the data except for GABA staining in nhr-67 mutant where Chi-square test was used; *p<0.05, **p<0.01 and ***p<0.001 compared to single mutants; for expression in RMEs statistics were performed only on the category 4RMEs)) (B) AVL, RIS and RMEs are affected in nhr-67 null mutants. nhr-67(ot795) animals are not viable and otEx5999 is an extrachromosomal array that contains copies of the wild-type nhr-67 locus as well as an array marker (unc-47prom::mChOpti). Animals that either contain or do not contain this array were scored for mutant phenotypes at the L1 stage. (C) lim-6 null mutant animals display defects in GABAergic neuron identity of AVL, RIS and DVB. (D) nhr-67 and lim-6 interact genetically in the AVL neuron. Expression of unc-25/GAD and unc-46/LAMP in AVL were scored at L1 in the different mutant background. (Chi-square test, **p<0.01 compared to single mutants) (E) ceh-10 mutant animals display defects in RMED GABAergic neuron identity. Expression of GABA pathway genes in null mutants (gm133) was scored as L1, when they arrest development. Viable gm127 hypomorphic mutants were scored as adults. Wild type data in panel C are re-iterated in panel E. (F) ceh-10 does not affect the generation of RMED as assessed by expression of a pan-neuronal marker in red but affects GABAergic fate as assessed by the absence of the unc-47/VGAT reporter in green. (A–F) (plain arrow head indicates a neuron presence and empty arrow head its absence) (Scale bar = 10 μm)
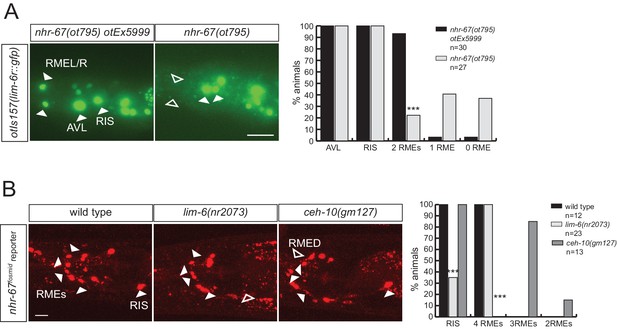
Regulatory relation of nhr-67 and other homeobox genes.
(A) lim-6 expression is regulated by nhr-67 in RMEL/R but not in RIS or AVL. lim-6::gfp reporter otIs157 (entire locus fused to gfp) was scored in a wild-type and nhr-67 null mutant background at the L1 stage. (B) nhr-67 fosmid expression is controlled by ceh-10 in RMED and by lim-6 in RIS. nhr-67::mChOpti rescuing fosmid reporter otEx3362 was scored in the wild-type background, lim-6 and ceh-10 mutant background at the adult stage. (A–B) (Scale bar = 10 μm, Fisher exact test, ***p<0.001, compared to wild type; when not noted, not significant)
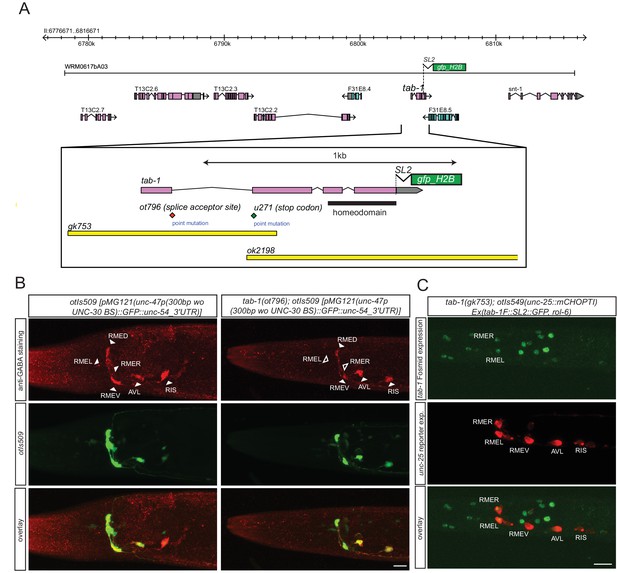
tab-1 is a homeodomain protein cooperating with nhr-67 to specify RMEL/R GABAergic identity.
(A) tab-1 locus, alleles and fosmid reporter. (B) tab-1 affects in RMEL/R unc-47 reporter expression (in green) and anti-GABA staining (in red). The unc-47 reporter used has a presumptive unc-30 binding site deleted, which diminishes the expression of this reporter in D-type neurons. The effects on these and other reporters are quantified in Table 5 (empty arrow head indicates loss of expression). (C) Expression pattern of the rescuing tab-1 fosmid reporter (in green) in a tab-1 mutant background with a unc-25/GAD gene reporter (in red). (B–C) (Scale bar = 10μm)
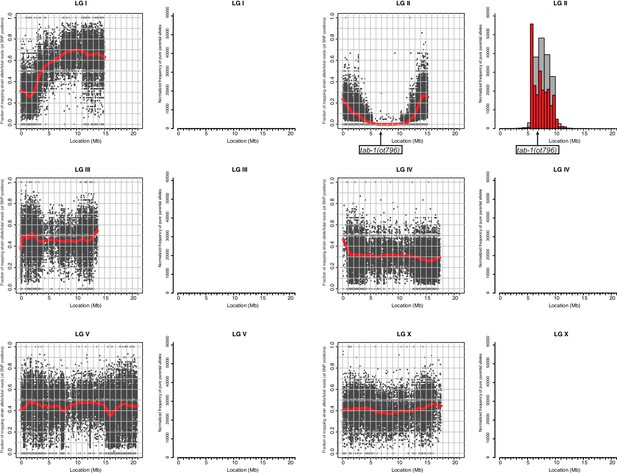
Whole genome sequencing data for tab-1 mutant identification.
A previously described mapping/sequencing approach was used to map the tab-1 mutant allele (Doitsidou et al., 2010). Data was analyzed using CloudMap (Minevich et al., 2012).
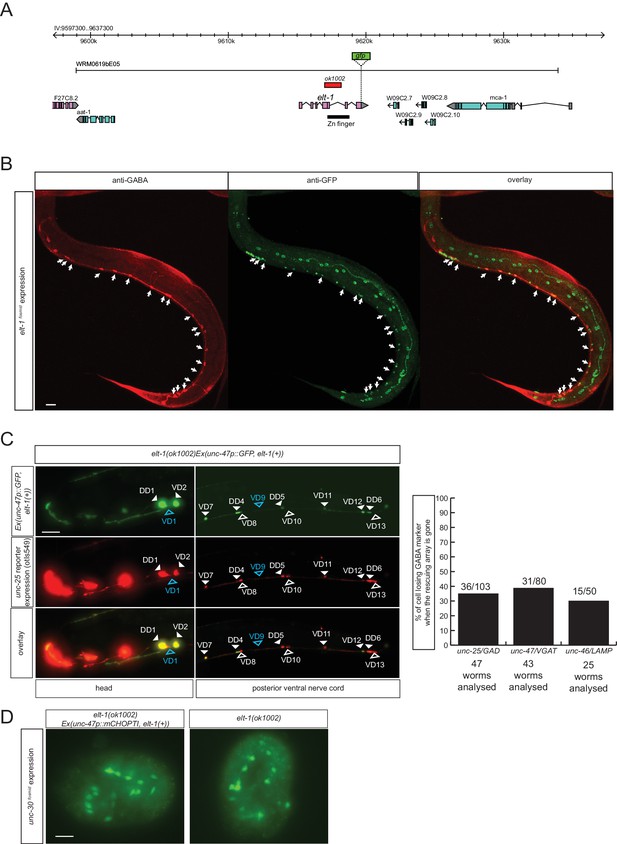
The GATA2/3 ortholog elt-1 controls GABAergic identity of D-type motor neurons.
(A) elt-1 locus and fosmid reporter used for expression analysis. The same, but untagged fosmid was used for mosaic analysis (panel C). (B) elt-1 is expressed in D-type motor neurons. The expression pattern of the elt-1 fosmid based reporter construct shown after immunostaining with anti-GFP in green and anti-GABA in red (C) Mosaic analysis of elt-1 function. elt-1 null mutants that carry a rescuing array, as well as an array marker that labels the presence of the array in GABA neurons (unc-47::gfp) are scored for unc-25::mChopti (otIs549) expression. Whenever no gfp signal is observed in a D-type motor neuron (empty arrow head), the expression of unc-25 prom::mChOpti is scored (white empty arrow head if present and blue empty arrow head if absent). Bar graph shows similar scoring done for the expression of unc-47/VGAT and unc-46/LAMP. (D) unc-30 fosmid reporter expression is not affected in elt-1 mutants, indicating that D-type motor neurons are born in the elt-1 mutants. (B–D) (Scale bar = 10 μm).
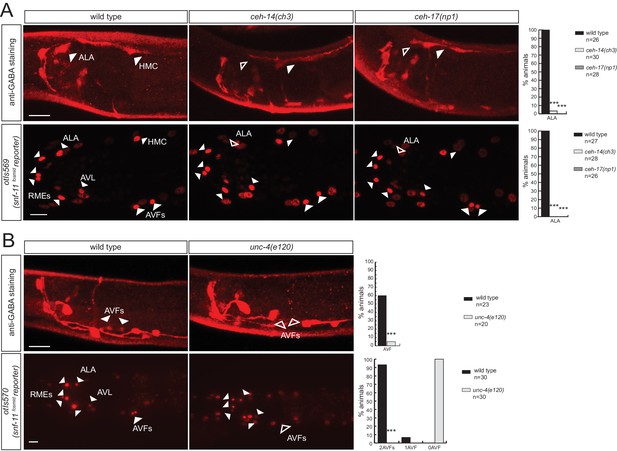
Homeobox genes controlling the identity of GABA uptake neurons.
(A) ceh-14 and ceh-17 specify the identity of the newly identified ALA GABAergic neuron, as assessed by GABA staining (upper panels) and snf-11 fosmid reporter expression (lower panels) (B)unc-4 affects GABA staining (upper panels) and snf-11 reporter gene expression (lower panels) in the AVF neuron. (A–C) plain arrow head indicates a neuron presence and empty arrow head its absence (Scale bar = 10 μm, Fisher exact test, ***p<0.001, compared to wild type; when not noted, not significant; for expression in AVFs statistics were performed only on the category 2AVFs)
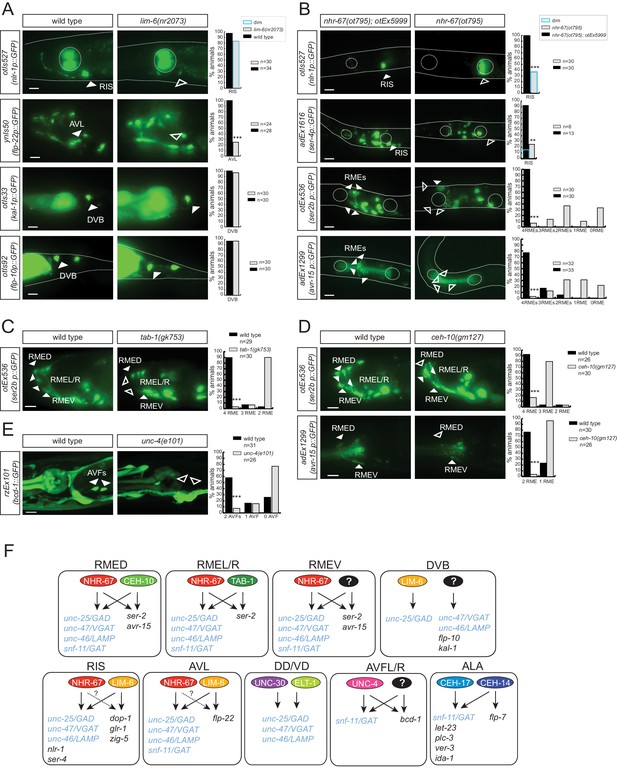
Transcription factors controlling GABAergic identity also control other cellular identity markers.
(A) lim-6 controls cell identity markers in AVL and RIS, but not DVB. (B) nhr-67 controls additional neuron identity markers of GABAergic neurons. nhr-67(ot795) animals are not viable and otEx5999 is an extrachromosomal array that contains copies of the wild-type nhr-67 locus as well as an array marker (unc-47prom::mChOpti). Animals that either contain this array or do not contain it were scored for mutant phenotypes at the L1 stage. (C) tab-1 controls the expression of RMEL/R identity markers. (D) ceh-10 controls the expression of RMED identity markers. (E) unc-4 controls the expression of the AVF identity marker bicd-1. (A–E) plain arrow head indicates a neuron presence and empty arrow head its absence (Scale bar = 10 μm; Fisher exact test, ***p<0.001, compared to wild type; when not noted, not significant; for expression in RMEs or AVFs statistics were performed only on the category 4RMEs or 2AVFs) (F) Summary of known gene regulatory programs in GABA producing neurons (GABA uptake neurons are not shown). ‘?’ indicates the regulatory relationship is just extrapolated from the effect of the transcription factor on GABA pathway genes. Already previously reported were the effect of lim-6 on non-GABA pathway genes in RIS and AVL, on unc-25 in DVB (Hobert et al., 1999; Tsalik et al., 2003), the effect of nhr-67 on unc-47 (Sarin et al., 2009), the effect of unc-30 on unc-25 and unc-47 (Eastman et al., 1999), the effect of ceh-14/ceh-17 on let-23, plc-3, ver-3, ida-1 and flp-7 (Van Buskirk and Sternberg, 2010).
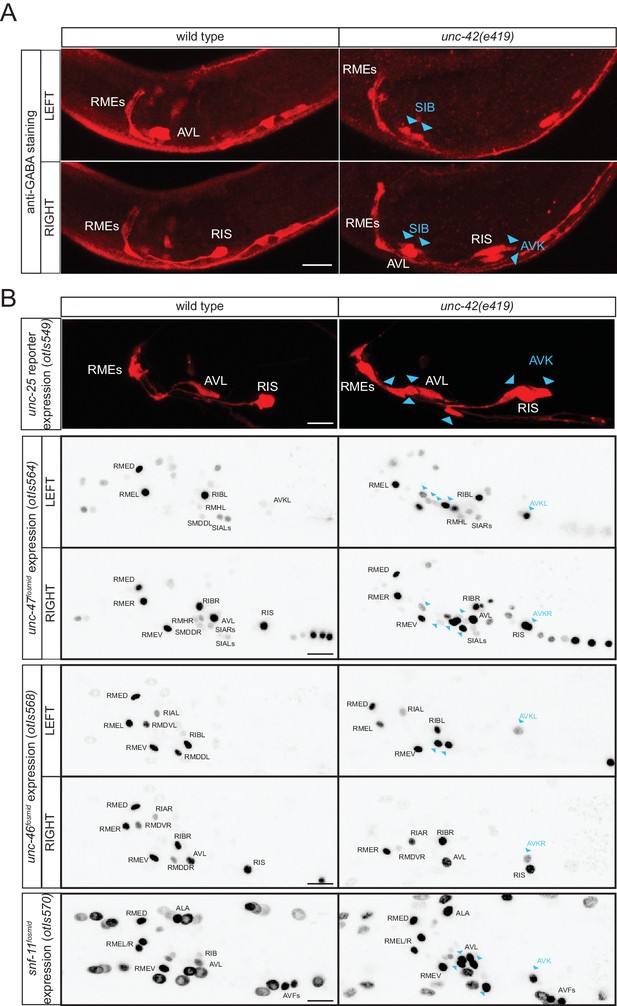
unc-42 represses GABAergic neuron identity.
(A) SIB and AVK switch to a GABA fate in unc-42 mutants. Anti-GABA staining in head neurons of wild-type and unc-42(e419) mutant. (B) GABAergic gene battery is turned on in additional neurons in unc-42 mutant animals. unc-25/GAD gene reporter expression (upper panel) and unc-47/VGAT, unc-46/LAMP and snf-11/GAT fosmid reporter expression in head neurons of wild-type and unc-42(e419) mutant animals. Fosmid reporter expression images are color-inverted. (A–B) Blue arrow heads point to ectopic expression. (Scale bar = 10 μm)
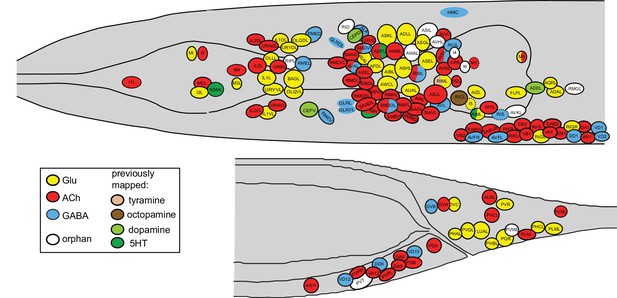
Neurotransmitter atlas.
The current status of the C. elegans neurotransmitter atlas is shown. Only the head and tail of the worm are shown (left view). The Glu and ACh maps come from (Pereira et al., 2015; Serrano-Saiz et al., 2013) and have been updated here with the GABA neuron analysis. See Supplementary file 1 for complete list. Dashed circle indicates neurons that are only present on the right side of the animal.
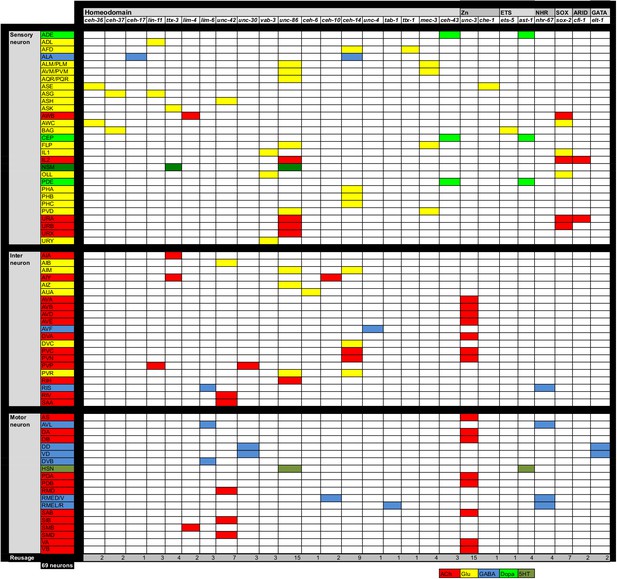
Regulatory map of neurotransmitter specification.
All neurons are shown for which neurotransmitter identity has been determined and for which a regulator for the respective neurotransmitter identity has been identified. Colored boxes indicate cells in which the indicated transcription factor affects neurotransmitter specification. Data are from this paper for GABA and from following papers for additional neurons: (Alqadah et al., 2015; Altun-Gultekin et al., 2001; Doitsidou et al., 2013; Flames and Hobert, 2009; Kratsios et al., 2015, 2012; Pereira et al., 2015; Serrano-Saiz et al., 2013; Sze et al., 2002; Vidal et al., 2015; Zhang et al., 2014)
Videos
Flyover movies ACh (red), Glu (mint), GABA (blue), monoaminergic (green) neurons.
This movie was generated by Chris Grove.
Tables
GABA-positive cells.
| Cells | anti-GABA | anti-GABA in snf-11(-) | anti-GABA in unc-47(-) | unc-25 knock-in reporter allele | unc-47 fosmid reporter | unc-46 fosmid reporter | snf-11 fosmid reporter | other fast neuro-transmitter | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Previously identified (Mclntire et al., 1993b) | Neuronal | RME | +++ | +++ | +++ | +++ | +++ | +++ | +++ | none |
| RIS | +++ | +++ | +++ | +++ | +++ | +++ | - | none | ||
| AVL | +++ | +++ | +++ | +++ | +++ | +++ | ++ | none | ||
| DVB | +++ | +++ | +++ | +++ | +++ | +++ | - | none | ||
| DD1-6 | +++ | +++ | +++ | +++ | +++ | +++ | - | none | ||
| VD1-13 | +++ | +++ | +++ | +++ | +++ | +++ | - | none | ||
| Newly identified | Neuronal | RIB | ++ | ++ | ++ | ++ | +++ | ++ | + | |
| SMDD/V | +/- | + | + | - | + | - | - | |||
| AVB | ++ | + | + | - | - | + | - | |||
| AVA | +/- | +/- | + | - | - | - | - | |||
| AVJ | ++ * | + | + | - | - | - | - | none | ||
| ALA (recycling) | ++ | - | - | - | + | - | +++ | none | ||
| AVF (clearance) | + | - | - | - | - | - | +++ | none | ||
| Non-Neuronal | GLR (non-neuronal) | + | - | ++ | - | - | + | ++ | none | |
| hmc (non-neuronal) | ++ | - | - | - | - | - | +++ | none | ||
| muscle | +++ | ++ | ++ | - | - | - | +++ | none | ||
Additional neurons in male | VD12 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | none | |
| EF1-4 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | none | ||
| R2A | + | + | + | - | - | - | - | |||
| R6A | ++ | ++ | ++ | - | + | - | - | |||
| R9B | + | + | + | - | - | +/- | - | none | ||
-
Additional sites of unc-46 and unc-47 expression are shown in Table 2. unc-25 and snf-11 are expressed exclusively in the cells shown here.
-
Color coding in column 3 (same as in Figure 2E):
-
Blue: 'conventional' GABA neurons that synthesize and synaptically release GABA.
-
Grey: non-conventional GABA neurons which acquire and release GABA by presently unknown means.
-
Purple: GABA uptake neurons that take up GABA via SNF-11 (based on snf-11 expression and snf-11-dependence of GABA staining).
-
Green: Non-neuronal GABA uptake cells.
-
+/- does not consistently stain in all animals. If present, staining is weak.
-
* Expression as strong as in RIB ('++') in larval stages, but intensity decreases in the adult.
Summary of unc-47/VGAT and unc-46/LAMP-expressing neurons in GABA-negative neurons. Expression in GABA-positive neurons is shown in Table 1. +/-, + and ++ represent relative expression levels. snf-11/GAT is expressed exclusively in the cells shown in Table 1.
| Cells | anti-GABA | unc-47 fosmid reporter | unc-46 fosmid reporter | snf-11 fosmid reporter | other neuro-transmitter |
|---|---|---|---|---|---|
| AIB | - | +/- | - | - | Glu |
| AIN | - | ++ | - | - | ACh |
| AIY | - | + | - | - | ACh |
| ALN | - | + | - | - | ACh |
| ASE | - | + | - | - | Glu |
| AUA | - | +/- | - | - | Glu |
| AVG | - | + | - | - | ACh |
| AVK | - | + | - | - | peptidergic |
| CEPV | - | + | - | - | dopamine |
| DVA | - | ++ | - | - | ACh |
| FLP | - | + | - | - | Glu |
| IL1 | - | +/- | - | - | Glu |
| OLL | - | +/- | - | - | Glu |
| PLN | - | + | - | - | ACh |
| PVT | - | ++ | - | - | none |
| RIA | - | +/- | ++ | - | Glu |
| RID | - | ++ | - | - | none |
| RIF | - | ++ | - | - | ACh |
| RIM | - | +/- | - | - | Glu |
| RIV | - | + | - | - | ACh |
| RMDD/V | - | - | ++ | - | ACh |
| RMH | - | ++ | - | - | ACh |
| SDQ | - | + | - | - | ACh |
| SIA | - | ++ | - | - | ACh |
| URX | - | ++ | - | - | ACh |
| URY | - | +/- | - | - | Glu |
| MI | - | ++ | - | - | Glu |
| M1 | - | + | - | - | ACh |
| M2 | - | + | + | - | ACh |
| M3 | - | + | - | - | Glu |
| M4 | - | ++ | - | - | ACh |
| I2 | - | ++ | - | - | Glu |
| I4 or I6 | - | ++ | - | - | none |
| I5 | - | +/- | - | - | Glu |
| NSM | - | + | - | - | 5HT |
Synaptic targets of GABA(+); UNC-47(+) neurons. Coloured box indicates that GABAergic output neuron synapses onto this target cell (<5 synapses [grey]; 5-20 synapses [orange]; >20 synapses [pink]) and genes in the boxes represent expression of ionotropic GABAA-type receptor subunits in the target cell (Figure 4). For a complete list of GABA receptor (+) neurons, see Table 4. Blue shading indicates that the target neurons is GABA(+). Wiring data is from www.wormwiring.org and includes all synapses observed. * = RMDL/R only.
| Targets | GABAergic output neurons | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| ALA | RIB | RIS | AVL | DVB | RME | SMD | DD/VD | ||
| muscle | exp-1, lgc-37, gab-1 | exp-1, lgc-37, gab-1 | unc-49, lgc-37, gab-1 | unc-49, lgc-37, gab-1 | unc-49, lgc-37, gab-1 | ||||
| sensory neuron(9 classes) | ASG | ||||||||
| ASJ | |||||||||
| BAG | |||||||||
| CEPD | |||||||||
| CEPV | |||||||||
| IL1 | |||||||||
| IL2 | lgc-38 | ||||||||
| PHC | |||||||||
| OLL | lgc-38 | lgc-38 | lgc-38 | ||||||
| URY | |||||||||
| inter-neuron (22 classes) | AIB | lgc-38 | lgc-38 | ||||||
| AIN | lgc-38 | ||||||||
| AIZ | lgc-37, lgc-38, gab-1 | lgc-37, lgc-38, gab-1 | |||||||
| ALN | |||||||||
| AUA | |||||||||
| AVA | lgc-37, gab-1 | lgc-37, gab-1 | |||||||
| AVB | |||||||||
| AVD | lgc-35 | lgc-35 | |||||||
| AVE | gab-1 | gab-1 | gab-1 | gab-1 | gab-1 | ||||
| AVK | |||||||||
| DVC | |||||||||
| PVP | |||||||||
| PVR | lgc-37, gab-1 | ||||||||
| PVW | lgc-37, gab-1 | ||||||||
| RIA | |||||||||
| RIB | |||||||||
| RIG | |||||||||
| RIH | |||||||||
| RIP | |||||||||
| RIS | |||||||||
| SAA | |||||||||
| Motor neuron (16 classes) | AVL | ||||||||
| DA | lgc-37, gab-1, lgc-35 | ||||||||
| DD | |||||||||
| DVB | |||||||||
| HSN | lgc-37, gab-1 | ||||||||
| PDA | exp-1, lgc-35 | ||||||||
| RIM | |||||||||
| RIV | lgc-38 | ||||||||
| RME | |||||||||
| RMD* | gab-1, lgc-37 | gab-1, lgc-37 | gab-1, lgc-37 | gab-1, lgc-37 | gab-1, lgc-37 | ||||
| RMH | |||||||||
| SAB | exp-1, lgc-37, gab-1, lgc-36 | ||||||||
| SIA | lgc-37, gab-1 | ||||||||
| SIB | |||||||||
| SMD | lgc-37, lgc-38, gab-1 | lgc-37, lgc-38, gab-1 | lgc-37, lgc-38, gab-1 | lgc-37, lgc-38, gab-1 | |||||
| VC | lgc-37, gab-1 | ||||||||
| VD | |||||||||
Summary of expression of GABAA-type receptors. gab-1, lgc-37, lgc-38 and lgc-36 expression is from this paper (Figure 4), exp-1 is from Beg and Jorgensen (2003), lgc-35 from Jobson et al. (2015) and unc-49 from Bamber et al. (1999). Cells that receive GABAergic synaptic input (as shown in wormwiring.org) are boxed blue. Others likely receive GABA spillover signals. For lgc-38, all expressing cells shown are observed with the 3.5 kb reporter fusion, except for OLL, which only expresses the 3.9 kb fusion; URA expresses both.
| cell type | gab-1 | lgc-37 | lgc-38 | lgc-36 | exp-1 | lgc-35 | unc-49 |
|---|---|---|---|---|---|---|---|
| ADE | |||||||
| AIB | |||||||
| AIM | |||||||
| AIN | |||||||
| AIY | |||||||
| AIZ | |||||||
| ALM | |||||||
| ASH | |||||||
| AVA | |||||||
| AVD | |||||||
| AVE | |||||||
| AVF | |||||||
| AVM | |||||||
| AS | |||||||
| BDU | |||||||
| DA | |||||||
| DB | |||||||
| DVA | |||||||
| FLP | |||||||
| HSN | |||||||
| IL2 | |||||||
| OLL | |||||||
| PDA | |||||||
| PLM | |||||||
| PLN | |||||||
| PVC | |||||||
| PVD | |||||||
| PVM | |||||||
| PVR | |||||||
| PVT | |||||||
| PVW | |||||||
| RID | |||||||
| RIV | |||||||
| RMD* | |||||||
| RMF | |||||||
| SABD | |||||||
| SABV | |||||||
| SDQR † | |||||||
| SIA | |||||||
| SMD | |||||||
| URA | |||||||
| URB | |||||||
| URX | |||||||
| VA | |||||||
| VB | |||||||
| VC | |||||||
| M3 | |||||||
| MC | |||||||
| MI | |||||||
| muscle |
-
* Expression is only observed in RMDL/R, not RMDD/V.
-
† Asymmetrically expressed in only SDQR, not SDQL.
Genetic characterization of tab-1 function in the RME neurons. Expression in RME can not always be unambiguously assigned to the dorsal, ventral, left or right RME neuron, hence only the total number of RME neurons was counted. otEx6804-6806 are tab-1-rescuing arrays (see Material and methods).
| marker | Genotype | Expression observed in: | ||||||
|---|---|---|---|---|---|---|---|---|
| 4 RMEs | 3 RMEs | 2 RMEs | 1 RME | 0 RME | n | |||
| unc-47prom::gfp (otIs509) | wild type | 100% | 0% | 0% | 0% | 0% | 31 | |
| ot796 | 0% | 26.7% | 70% | 3.3% | 0% | 30 | ||
| unc-47prom::gfp (oxIs12) | wild type | 100% | 0% | 0% | 0% | 0% | 30 | |
| gk753 | 16.7% | 30% | 53.3% | 3.3% | 0% | 30 | ||
| unc-25prom::gfp (otIs549) | wild type | 100% | 0% | 0% | 0% | 0% | 30 | |
| ot796/+ | 100% | 0% | 0% | 0% | 0% | 22 | ||
| ot796 | 0% | 23.3% | 76.7% | 0% | 0% | 30 | ||
| gk753 | 0% | 20% | 80% | 0% | 0% | 30 | ||
| ot796 otIs549/+ | 6.7% | 36.7% | 56.6% | 0% | 0% | 30 | ||
| ot796/ok2198 | 0% | 42.8% | 57.2% | 0% | 0% | 28 | ||
| ot796/u271 | 3.3% | 36.7% | 60% | 0% | 0% | 30 | ||
| ot796/gk753 | 3.3% | 36.7% | 60% | 0% | 0% | 30 | ||
unc-46prom::gfp(otIs575) | wild type | 93.3% | 6.7% | 0% | 0% | 0% | 30 | |
| gk753 | 13.3% | 36.7% | 46.7% | 3.3% | 0% | 30 | ||
| anti-GABA staining | wild type | 70% | 16.7% | 10% | 0% | 3.3% | 30 | |
| ot796 | 10% | 20% | 63.4% | 3.3% | 3.3% | 30 | ||
tab-1(gk753); otIs549; Ex[tab-1(+),ttx-3::gfp] | without otEx6804 | 13.3% | 16.7% | 70% | 0% | 0% | 30 | |
| with otEx6804 | 80% | 8% | 12% | 0% | 0% | 25 | ||
| without otEx6805 | 5% | 25% | 65% | 0% | 5% | 20 | ||
| with otEx6805 | 41.9% | 9.7% | 48.4% | 0% | 0% | 31 | ||
| without otEx6806 | 5.6% | 16.7% | 77.8% | 0% | 0% | 18 | ||
| with otEx6806 | 66.7% | 10% | 23.3% | 0% | 0% | 30 | ||
Additional files
-
Supplementary file 1
A list of all C. elegans neuron classes and their assigned neurotransmitter identities.
Data from (Chase and Koelle, 2007; Pereira et al., 2015; Serrano-Saiz et al., 2013).
- https://doi.org/10.7554/eLife.17686.022





