Formation of polarity convergences underlying shoot outgrowths
Figures
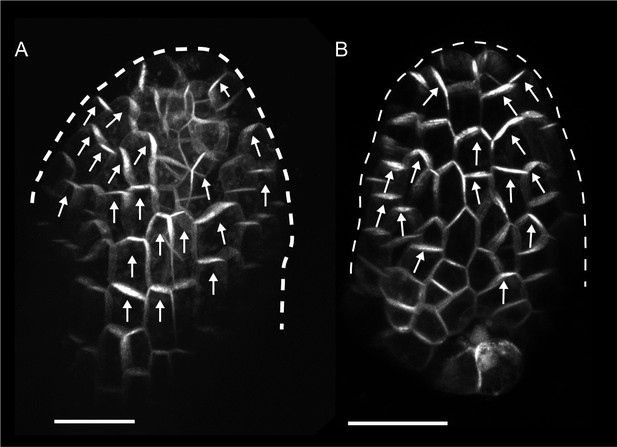
PIN1::PIN1:GFP expression in young WT and kan1kan2 leaf primordia.
(A) WT primordium of leaf 1, showing abaxial epidermis (a total of 10 leaves were imaged over 2 separate experiments). (B) As for A, but for a kan1kan2 primordium (a total of 15 leaves were imaged over three separate experiments). White arrows indicate inferred polarities. Dashed white lines indicate leaf outlines. Scale bars = 20 µm.
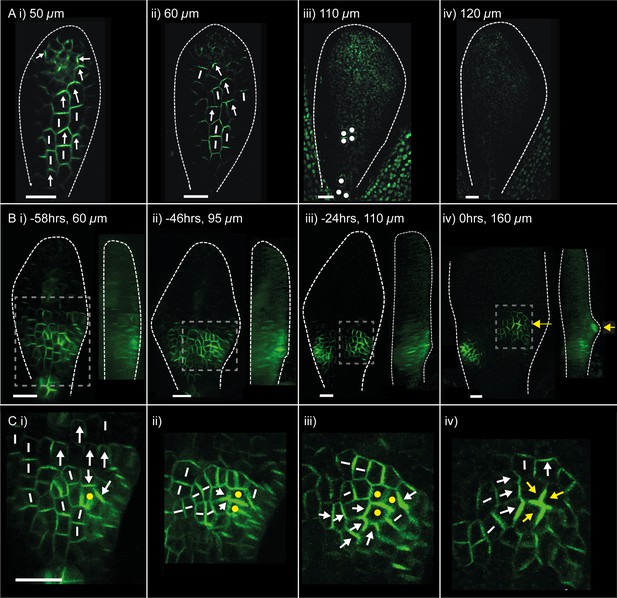
PIN1::PIN1:GFP polarity patterns in WT and kan1kan2 leaf development.
Confocal images of the PIN1::PIN1:GFP marker in the abaxial epidermis of the same WT leaf primordium imaged over a period of 2 days. Approximate leaf widths (measured from projections of the z stacks) are given above each image. Times from the beginning of the experiment are: i) 0 hrs, ii) 12 hrs, iii) 36 hrs, iv) 48 hrs. Cells indicated by white dots in iii) retain detectable expression of the marker for longer than other cells. This time-lapse data is supported by snapshot images taken for at least five leaves at each developmental stage in a separate experiment. (B) Confocal images of the PIN1::PIN1:GFP marker in the abaxial epidermis of a kan1kan2 leaf primordium, prior to and during the emergence of an ectopic outgrowth. For each time point, a surface view of the abaxial epidermis (left) and a side view of a 3D rendering of the confocal z-stack (right) are shown. The side views allow the emergence of the outgrowth to be monitored. The time relative to when an outgrowth could clearly be observed and the estimated leaf width are given above each image. Yellow arrows in iv) indicate an ectopic centre of PIN1 polarity convergence at the tip of an emerging outgrowth. (C) Magnified images of the regions outlined by the dashed grey rectangles in B, showing the development of the centre of PIN1 polarity convergence at the tip of the emerging outgrowth. Yellow dots, arrows and lines indicate the cells that form the centre of convergence in iv). Arrows indicate inferred PIN1 polarities and lines indicate inferred axes of PIN1 distributions. Scale bars = 20 µm. The scale bar in C i) applies to all panels in C. This data is representative of tracking three out of three kan1kan2 leaves that developed ectopic outgrowths, each in a separate experiment, and with snapshot images from at least three leaves at each developmental stage (in another separate experiment).
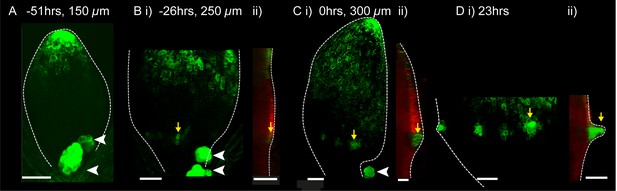
DR5::GFP expression in a kan1kan2 leaf during outgrowth development.
Confocal images of DR5::GFP in the same kan1kan2 leaf imaged over a period of 3 days. Times relative to the first observation of an outgrowth, and leaf widths, are given above images. B ii, C ii and D ii show optical sections through 3D renderings of confocal z-stacks. DR5::GFP (green) and auto-fluorescence plus CUC2::RFP (red) channels are shown (CUC2::RFP is used to help show the leaf outline). Yellow arrows in D indicate the region of DR5::GFP activity at the tip of an outgrowth and yellow arrows in B and C indicate the same DR5::GFP expressing cells tracked back in time prior to outgrowth emergence. White arrow heads indicate high DR5::GFP signal in stipules. Scale bars = 50 µm. This data is representative of tracking 4 out of 4 kan1kan2 leaves that developed outgrowths (in two experiments), and of snapshot data taken for at least 3 leaves at each of the developmental stages shown (in another separate experiment).

Leaf phenotype of the kan1kan2cuc2 mutant.
(A) Whole kan1kan2 plant. (B) OPT images of a kan1kan2 leaf showing abaxial leaf surface. (C) Whole kan1kan2cuc2 plant. (D) OPT images of a kan1kan2cuc2 leaf. Scale bars in A and C = 1 cm, scale bars in B and D = 1 mm. See Figure 4—figure supplement 1 for more details of the kan1kan2cuc2 phenotype.

Abaxial ridges and serrations produced in kan1kan2cuc2 mutants.
(A) Example of a kan1kan2cuc2 leaf with an abaxial ridge-like thickening (indicated by blue arrow). (B) Example of a kan1kan2cuc2 leaf with serrations. Scale bars = 1 mm.
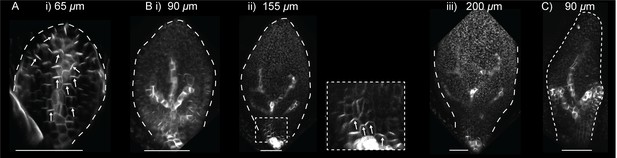
PIN1::PIN1:GFP in leaf one of the kan1kan2cuc2 mutant.
(A) Confocal image of PIN1::PIN1:GFP in the abaxial epidermis of the first leaf primordium of a kan1kan2cuc2 mutant. (B) Time-lapse confocal images of the abaxial side of the first leaf of a kan1kan2cuc2 mutant, taken at successive time points. Bi, ii and iii are maximum intensity projections of the abaxial side of the leaf, and therefore signal from both epidermal and sub-epidermal cell layers is combined. The data set shown here is representative of that obtained by tracking three kan1kan2cuc2PIN1::PIN1:GFP leaves (from three different seedlings), and of snapshot images of 10 kan1kan2cuc2 first leaf primordia taken at each of the developmental stages shown. (C) Maximum intensity projection of the abaxial side of leaf 5 of a kan1kan2cuc2 mutant, showing an example of epidermal centres of PIN1 convergence (on left and right sides of the leaf). Scale bars = 50 μm.
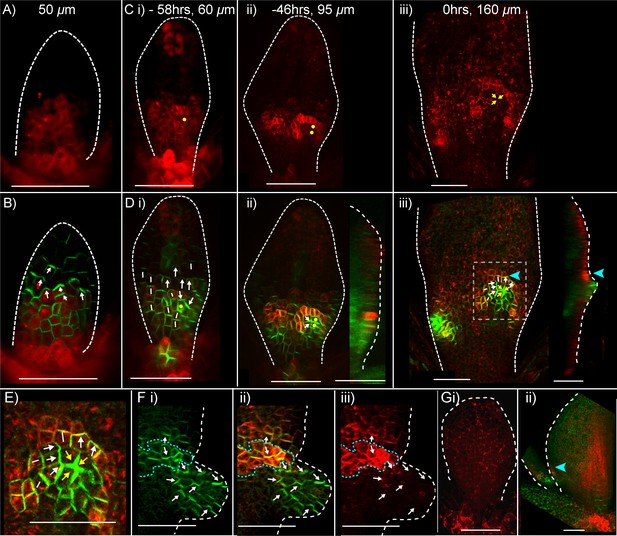
Expression of CUC2::RFPer and PIN1::PIN1:GFP during kan1kan2 outgrowth development.
(A) Confocal images of a CUC2::RFPer reporter in the abaxial epidermis of a young kan1kan2 first leaf primordium. (B) Combined CUC2::RFPer (red) and PIN1::PIN1:GFP confocal channels for the same leaf as in A. White arrows indicate inferred polarity orientations. (C) and (D) Time-lapse imaging of CUC2::RFP and PIN1::PIN1:GFP in the abaxial epidermis of the first leaf of another seedling from that in A and B, over 58 hr prior to outgrowth development. C: CUC2::RFPer, D: combined channels, as in B. Times relative to outgrowth emergence and approximate leaf widths are given above each image. The right-hand images in D ii and iii show optical sections through 3D renderings of the confocal z-stacks at the position of the centre of convergence. White and yellow arrows indicate inferred PIN1 polarity orientations. Yellow dots and arrows indicate the cells that gave rise to the outgrowth tip in C iii and D iii. Blue arrow heads in D iii indicate elevated CUC2::RFPer expression distal to a centre of PIN1 convergence and at the base of the emerging outgrowth on its distal side. Data is representative of that obtained by tracking outgrowth development in five kan1kan2 CUC2::RFPEer PIN1::PIN1:GFP leaves, in three separate experiments. (E) Zoomed-in image of the region outlined by the dashed rectangle in D iii, showing PIN1 polarities within and surrounding the domain of elevated CUC2::RFPer expression. (F) Confocal images of CUC2::RFPer (red) and PIN1::PIN1:GFP (green) associated with an ectopic kan1kan2 outgrowth at a later stage of development to that in C–E. Dotted blue line outlines the domain with elevated CUC2::RFPer expression at the distal boundary between an outgrowth and the main lamina. (G) CUC2::RFPer expression in two WT first leaf primordia (ii also shows PIN::PIN1:GFP in green). Blue arrow in ii indicates a site of elevated CUC2::RFPer expression in the leaf margin. Red signal in the centre of the lamina in ii) is non-ER localised and therefore due to auto-fluorescence. Data is representative of of that obtained by imaging nine leaves at each of the developmental stages shown, across two separate experiments. Scale bars = 50 μm. Dashed white lines indicate leaf outlines.
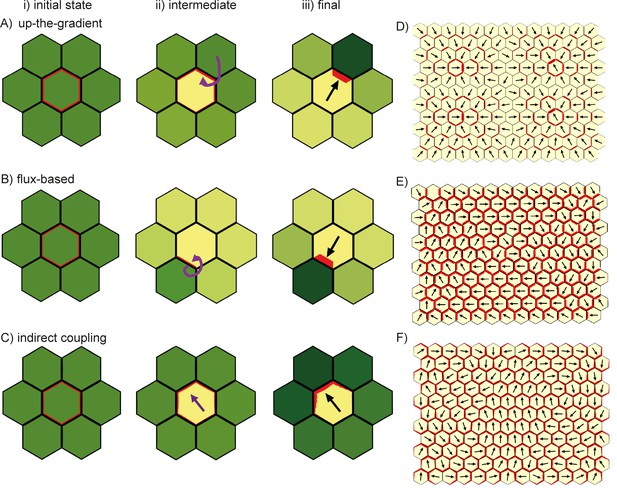
Comparison of basic behaviours of indirect coupling, flux-based and up-the-gradient models.
(A–C) Investigating the ability of cells to polarise in the absence of pre-established external asymmetries or polarisable neighbours for up-the-gradient (A), flux-based (B) and indirect coupling (C) models. Panels i) show the initial state of the simulations. In each simulation, all cells initially have the same auxin concentration (indicated by the intensity of green, where darker green indicates a higher concentration). In the case of up-the-gradient or flux-based models (A, B), noise is present in the initial levels of PIN1 (shown by red lines at cell edges). In the case of indirect coupling (C), a single central cell has noise in the levels of A* and B*. Panels ii) show intermediate states of the simulations, and panels iii) show the final states. In this and subsequent simulations, PIN1 polarisation is indicated by black arrows, where the arrow points towards the region of the cell with highest PIN1. (A) For the up-the-gradient model, if a given cell edge has slightly elevated PIN, this causes the juxtaposed neighbour to have an increased auxin concentration. This increased auxin concentration in the neighbour feeds-back to cause increased PIN1 recruitment to the given cell edge (purple arrow in ii). (B) For the flux-based model, a cell edge with a slightly elevated PIN1 concentration causes an increased auxin efflux rate. This increased auxin efflux feeds-back to cause an increased recruitment of PIN1 to the given cell edge (purple arrow in ii). (C) For the indirect coupling model, polarity is generated independently from PIN1 and auxin through A* and B* polarity components (purple arrow in ii) which in turn causes polarisation of PIN1. (D–F) Behaviour of up-the-gradient (D), flux-based (E) and indirect coupling (F) models for 2D arrays of polarisable cells in the absence of pre-established asymmetries. D and E were initialised with noise in the initial concentrations of auxin and F was initialised with noise in the levels A* and B* in the membrane. Details of all simulations can be found in the Materials and methods. See Figure 7—figure supplement 1 for a further comparison of the requirements for polarisation.
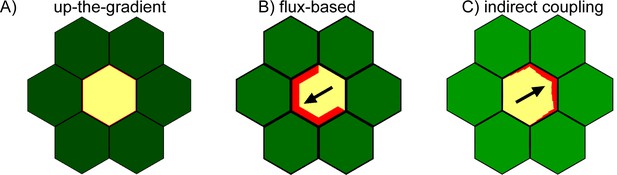
Comparison of models’ ability to generate polarity for a cell surrounded by a fixed environment.
In each case the central cell is initialized with noise in the concentrations of PIN at each cell edge and all seven cells have the same auxin concentration. The auxin concentration in each outside neighbor of the central cell is fixed throughout the simulation so that the central cell cannot induce asymmetries in its environment.
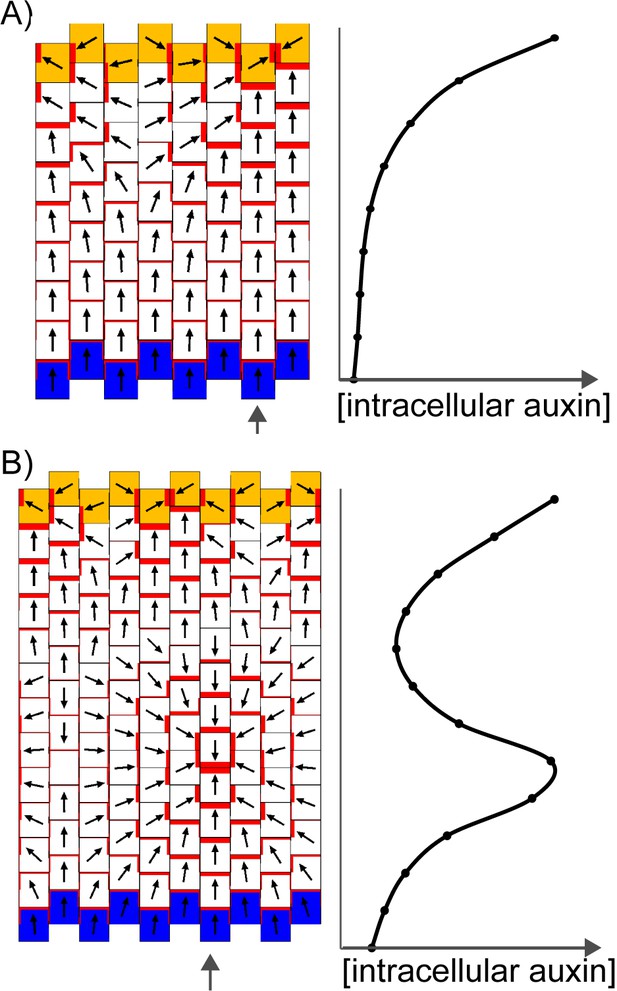
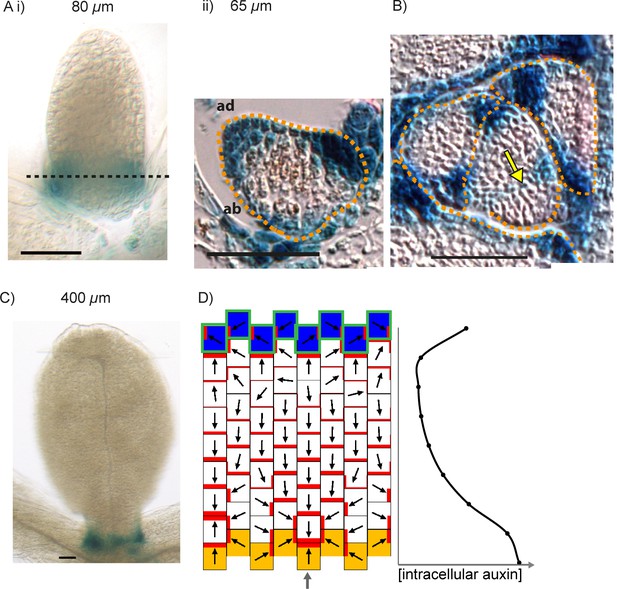
Formation of a proximo-distal polarity field and centres of convergence in the convergent alignment model.
(A) Formation of a proximo-distal polarity field due to the presence of an elevated initial auxin concentration at the leaf tip (orange cells) and an elevated rate of auxin removal from the leaf base (blue cells). The graph shows the concentration of intracellular auxin for each cell in the column marked with the grey arrow. (B) As for A, but for a larger tissue. A centre of polarity convergence forms within the proximal half of the lamina.
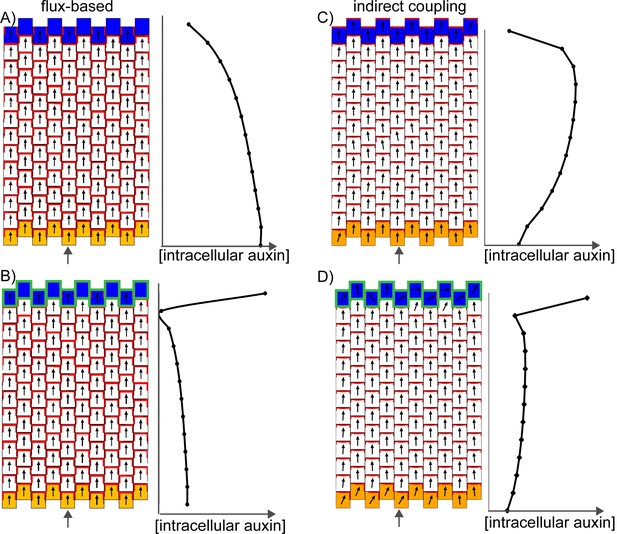
Formation of a proximo-distal polarity field in the flux-based and indirect coupling models.
(A) Formation of a proximo-distal polarity field in the flux-based model due to the presence of elevated auxin biosynthesis at the leaf base (orange cells) and an elevated rate of auxin removal from the leaf tip (blue cells). The graph on the right shows the concentration of intracellular auxin for the column marked with the grey arrow. (B) As for A, but with elevated auxin import (green cell outlines) at the distal end of the tissue. (C) As for A, but for the indirect coupling model. (D) As for B, but for the indirect coupling model.
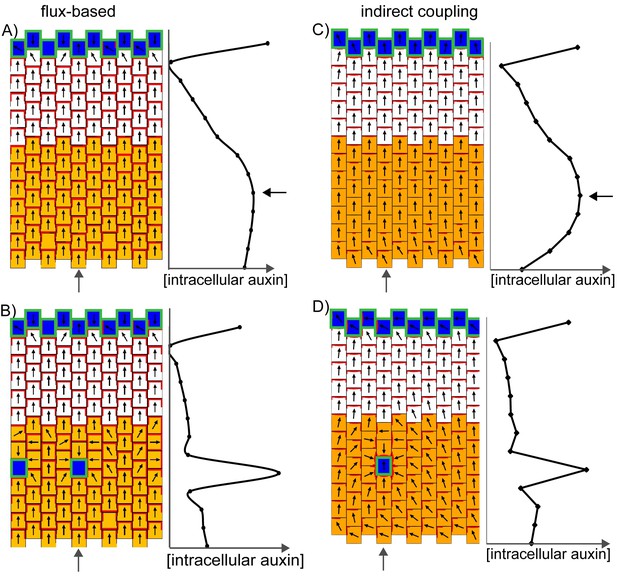
Formation of centres of polarity convergence in the flux-based and indirect coupling models.
(A) A proximo-distal polarity field is initially established with the flux based model due to the presence of elevated auxin biosynthesis in the proximal half of the leaf (orange cells) and elevated rates of auxin import and removal from the leaf tip (blue cells with green outlines). The graph on the right shows the concentration of intracellular auxin for the column marked with the grey arrow. Note that there is a broad peak of intracellular auxin concentration within the proximal domain of elevated auxin synthesis (black arrow). (B) Subsequent stage of the simulation in A. If it is assumed that intracellular auxin above a threshold concentration induces elevated auxin import and removal, one or more cells within the proximal domain of elevated auxin biosynthesis are induced to have elevated levels of import and removal. Neighbouring cells reorient their polarity to point towards the high import cells, which accumulate elevated levels of intracellular auxin. (C) As for A), but for the indirect coupling model. (D) As for B), but for the indirect coupling model. See Figure 10—figure supplement 1 for an alternative version of the indirect coupling model which incorporates D6-kinase like activity.
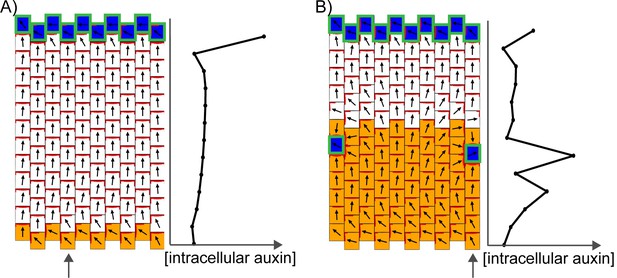
Incorporation of a D6 protein kinase-like activity into the indirect coupling model.
D6 kinase activity was added into simulations of proximodistal polarity establishment (A), (compare with Figure 9D) and convergence formation (B) (compare with Figure 10D). We assume that D6 protein kinase localizes to the A* end of the cell and locally enhances the export of auxin by PIN1. Since A* determines the distribution of PIN1 in the simple form of the indirect coupling model, this addition of D6-kinase-regulated PIN1 activity where A* is present makes little difference to the model’s behaviour.
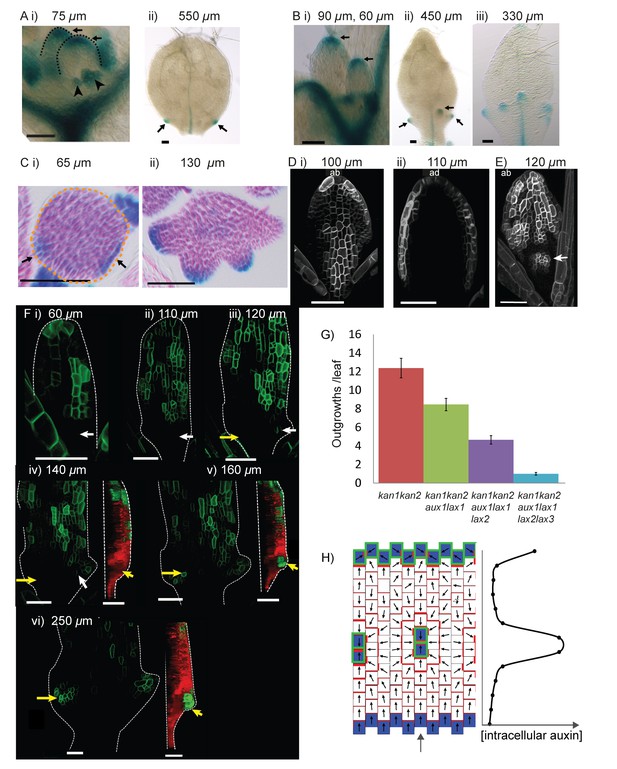
Expression of LAX1::GUS and AUX1::AUX1:YFP in WT and kan1kan2 leaves.
(A) Expression pattern of LAX1::GUS in WT leaves one and two. LAX1::GUS was expressed at the tips of developing primordia (arrows in (i), black dashed lines indicate leaf outlines, arrow heads indicate stipules) (a total of 6 plants (with 12 young leaves) were imaged across two separate experiments) and at the tips of serrations (ii) (arrows indicate serrations) (a total of 9 plants (18 leaves) were imaged across three separate experiments). Leaf widths are given above images. (B) Expression pattern of LAX1::GUS in kan1kan2 leaves. LAX1::GUS was expressed at the tips of primordia (i) (a total of 7 plants (14 leaves) were imaged across two separate experiments) and at the tips of outgrowths (ii and iii) (arrows) (20 out of 21 leaves imaged across three separate experiments). ii) shows leaf 1, iii) shows leaf 3. (C) Transverse sections through GUS stained kan1kan2 LAX1::GUS seedlings, showing points of LAX1::GUS expression before outgrowths have emerged (black arrows in i) (data supported by serial sections of 3 other leaves at similar developmental stages in a separate experiment) and at the tips of developing outgrowths (ii) (a total of 8 leaves were imaged across two separate experiments). Dashed orange line in i) indicates leaf outline. (D) AUX1::AUX1:YFP expression in leaf 1 of WT, showing abaxial surface (i) (a total of 15 leaves were imaged across two separate experiments) and adaxial surface (ii) (a total of 4 leaves were imaged in two separate experiments) of two different leaves. (E) AUX1::AUX1:YFP expression in leaf 1 of a kan1kan2 mutant, showing abaxial surface. Arrow points to the tip of an emerging outgrowth with locally elevated AUX1::AUX1:YFP signal (a total of 9 leaves were imaged, across three different experiments). (F) Time-lapse confocal imaging of AUX1::AUX1:YFP in the abaxial epidermis of the first leaf of a kan1kan2 seedling. White arrows mark the positions of cells that eventually gave rise to the AUX1::AUX1:YFP expressing cells in the developing serration on the right side of the leaf. Yellow arrows mark the positions of cells which eventually gave rise to the tip of the ectopic outgrowth on the left side of the leaf. Red and green images in iv), v) and vi) show side views, from the left hand side of the leaf (showing that the outgrowth emerged at the time point shown in v). Red shows auto-fluorescence and a CUC2::RFP marker (used to show the leaf contours), and green shows AUX1::AUX1:YFP signal. Times from the beginning of the experiment at which images were taken are: i) 0 hrs, ii) 22 hr 40 min, iii) 31 hr 10 min, iv) 46 hr 40 min, v) 55 hr 40 min, vi) 74 hr 40 min. Data is consistent with tracking experiments performed for two other kan1kan2 leaves that developed ectopic outgrowths, and with snapshot images of at least 6 leaves before and after outgrowth emergence. (G) Mean number of outgrowths (+/- standard error of the mean) in rosette leaves of kan1kan2 plants carrying mutant alleles of AUX/LAX genes. n numbers are: kan1kan2: 84 leaves from 10 plants, kan1kan2aux1lax1: 110 leaves from 10 plants, kan1kan2aux1lax1lax2: 136 leaves from 10 plants, kan1kan2aux1lax1lax2lax3:200 leaves from 10 plants. (H) Convergent alignment model where elevated intracellular auxin causes elevated rates of auxin import and removal. The model is initialised with elevated auxin degradation at the leaf base (blue cells) and an elevated rate of auxin import and degradation at the leaf tip (blue cells with green outlines). Centres of convergence form within the proximal half of the lamina and when the intracellular auxin concentration exceeds a threshold level, an elevated rate of auxin import and removal is induced (blue cells with green outlines). Scale bars = 50 µm. See Figure 11—figure supplement 1 for LAX2::GUS and LAX3::GUS expression.
-
Figure 11—source data 1
Counts of outgrowths used to generate Figure 11G.
- https://doi.org/10.7554/eLife.18165.017

Expression patterns of LAX2::GUS and LAX3::GUS.
(A) Expression of LAX2::GUS in a kan1kan2+/- leaf, showing expression in leaf vascular tissue. Scale bar = 100 μm. (B) Expression of LAX3::GUS in a WT seedling, showing expression is absent from leaves, but present in the hypocotyl and root. Scale bar = 1 mm (C) Expression of LAX3::GUS in a kan1kan2 seedling. Scale bars = 100 μm.
LAX1 and AUX1 expression in kan1kan2cuc2 mutants.
(A) Expression of LAX1::GUS in the abaxial surface of leaf 1 of a kan1kan2cuc2 mutant. Data representative of images from ten seedlings in two separate experiments. (B) AUX1::AUX1:YFP expression in leaf 3 and 4 of a kan1kan2cuc2 mutant. Images representative of those taken from fifteen seedlings in two separate experiments. (C) CUC2::RFPer and AUX1::AUX1:YFP expression in leaf 3 of a kan1kan2 mutant leaf i) AUX1::AUX1:YFP, ii) CUC2::RFPer (red) and AUX1::AUX1:YFP (green), iii) CUC2::RFPer. White arrows indicate region of low AUX1::AUX1:YFP and elevated CUC2::RFPer expression at the distal base of developing outgrowths. Images representative of those taken from six seedlings in two experiments. Approximate leaf widths are given above each image. Scale bars = 50 μm.
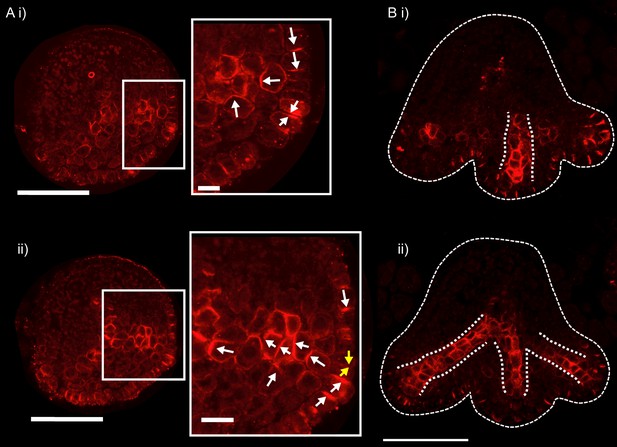
PIN1 immuno-localisation in transverse cross-sections of kan1kan2 leaves.
(A) PIN1 immuno-localisation in a kan1kan2 leaf before outgrowth emergence. i) and ii) are consecutive sections through the tissue (each section is 8 µm thick). A centre of PIN1 polarity convergence can be seen in i) and the site of this convergence is shown with yellow arrows in ii). Arrows show the inferred directions of PIN1 polarity. The adaxial side of the leaf is at the top of the image. 4 leaves with convergences were imaged across two separate experiments. (B) PIN1 immuno-localisation in a kan1kan2 leaf in which outgrowths have begun to emerge. Dotted white lines indicate the strands of cells with elevated levels of PIN1 and the leaf outline. 3 leaves with emerging outgrowths were imaged across two separate experiments. All scale bars = 50 μm, except those in the zoomed in panels in A, which are 10 μm.
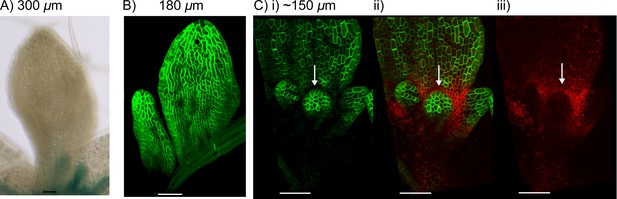
YUCCA1::GUS expression in WT leaves.
(A) Expression of YUC1::GUS in a WT leaf 1 primordium. i) abaxial epidermis (10 plants were imaged across two separate experiments) ii) transverse cross-sectional view of leaf 1 at a similar developmental stage (adaxial (ad) is towards the top) at the position marked by the dashed black line in i). Dashed orange line indicates leaf outline. Data consistent with serial sections taken through 3 other leaves. (B) Transverse cross-sectional view of a YUC1::GUS expressing wild-type vegetative meristem. Yellow arrow with black outline indicates region of YUC1::GUS expression at the boundary of an emerging primordium. Dashed orange lines indicate leaf primordia outlines. Data consistent with sections from 3 meristems, across two separate experiments. (C) Expression of YUC1::GUS at a later stage of WT leaf development. Scale bars = 50 µm. 9 leaves imaged across two separate experiments. (D) In the presence of elevated auxin production in the proximal row of cells (orange), and elevated auxin import and removal in the distal row of cells (blue cells with green outline), the convergent alignment model generates a divergent polarity field, with proximally oriented polarity in the proximal half of the tissue, and distally oriented polarity in the distal half.
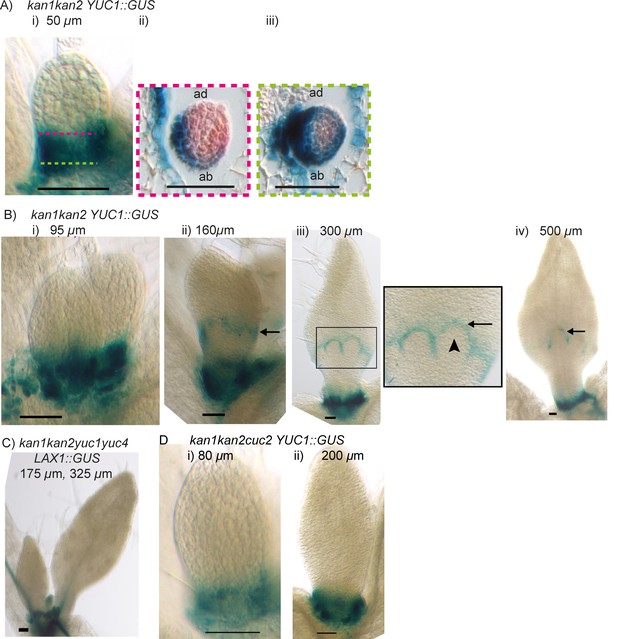
Expression of YUCCA1::GUS in kan1kan2 leaves.
(A) Expression of YUC1::GUS in kan1kan2 leaf one primordia. i) abaxial epidermis (a total of 15 leaves imaged, in three separate experiments), ii) transverse cross-sectional view of leaf 1 at a similar developmental stage, at an approximate position along the proximo-distal axis marked with the pink dashed line in i, iii) transverse cross section through the same leaf as shown in ii), at the approximate position marked with the green dashed line in i. Data consistent with serial sections for 3 other kan1kan2 leaves. (B) Expression of YUC1::GUS in kan1kan2 mutant leaf 1 primordia at progressive stages of development. Black arrows indicate the band of YUC1::GUS expression, which is at the distal base of outgrowths in iii) and iv). Arrow heads indicate an outgrowth tip which does not express YUC1::GUS. At least 8 leaves were imaged for each developmental stage, across two experiments. (C) Expression of LAX1::GUS in first two leaves of a kan1kan2yuc1yuc4 seedling (a total of 20 leaves were imaged in three separate experiments). (D) Expression of YUC1::GUS in leaf one of kan1kan2cuc2 mutant seedlings (15 seedlings were imaged at each developmental stage across three separate experiments). Scale bars = 50 µm.
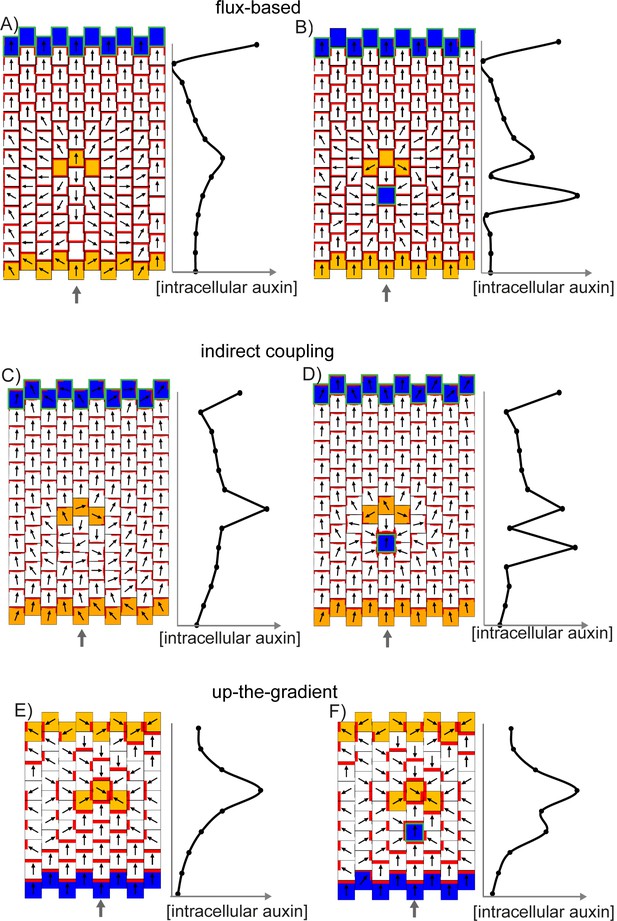
Effect of a band of locally elevated auxin biosynthesis on flux-based, indirect coupling and up-the-gradient models.
(A) Flux-based model. A proximodistal polarity field is initially established due to elevated auxin production at the leaf base (orange cells) and elevated auxin import and removal at the leaf tip (blue cells with green outline). After distally oriented polarities are established, 3 cells are given an elevated rate of auxin biosynthesis (orange cells), causing polarities to diverge away from this region. Graph shows intracellular auxin concentrations for the column of cells marked with the grey arrow. (B) As for A, but with a cell with elevated auxin import and removal (blue cell with green outline) on the proximal side of the band with elevated auxin synthesis. (C) As for A, but for the indirect coupling model. (D) As for B, but for the indirect coupling model. (E) Up the gradient model. A proximo-distal polarity field is initially established through elevated auxin removal at the leaf base (blue cells) and an initially elevated auxin concentration at the leaf tip (same initial conditions as Figure 8A). Three cells (isolated orange cells) are then given an elevated auxin biosynthesis rate, causing polarities to orient towards them. (F) As for E, but a cell with elevated auxin import and removal is also added on the proximal side of the band with elevated auxin synthesis.
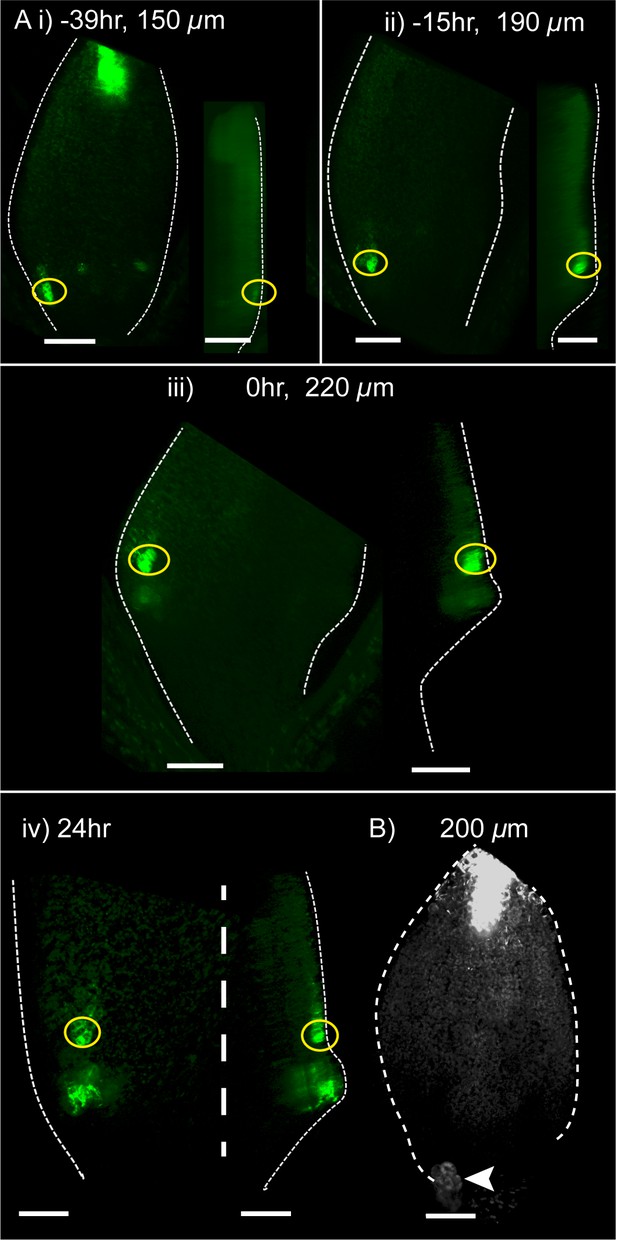
Time-lapse imaging of YUC4::GFP in kan1kan2 leaves.
(A) Confocal images of YUC4::GFP in the same kan1kan2 leaf imaged over a period of 3 days. Times relative to outgrowth emergence, and leaf widths, are given above images. Yellow circles indicate cells located distal to the outgrowth after its emergence, and their ancestors prior to outgrowth emergence. Note that these cells express YUC4::GFP before and after outgrowth emergence, but the region of expression at the outgrowth tip only appears following outgrowth emergence. Data is representative of tracking 5 out of 5 kan1kan2 leaves that developed ectopic outgrowths across two experiments. (B) YUC4::GFP expression in leaf one of a kan1kan2cuc2 mutant. Data representative of 13 leaves imaged in two separate experiments. Arrow head indicates stipule. Scale bars = 50 µm.
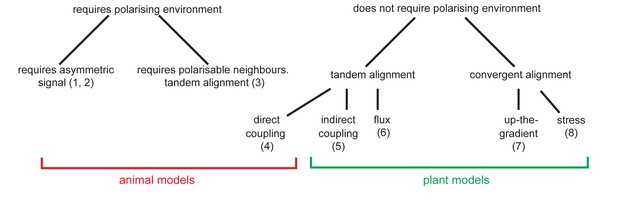
Classification of plant and animal polarity models.
Models of plant and animal epidermal polarity may be classified according to whether they require the presence of pre-established asymmetries or polarisable neighbours (referred to as a polarising environment) for cell polarity to arise. All plant models tested can generate cell polarity without a polarising environment, whilst animal models except the direct coupling model ([4] [Abley et al., 2013]) either require an asymmetric signal ([1] [Le Garrec et al., 2006] [2] [Amonlirdviman, 2005]) or polarisable neighbours ([3] [Burak and Shraiman, 2009]) for cell polarity to arise. A further classification is made according to the behaviours of groups of cells in the absence of pre-established asymmetries. In this context, the indirect coupling ([5] [Abley et al., 2013]) and flux-based ([6] [Mitchison, 1980; Rolland-Lagan and Prusinkiewicz, 2005; Stoma et al., 2008]) models of plant polarity generate swirled patterns of polarity with tandem alignments between cells. In contrast, the up-the-gradient model ([7] [Bilsborough et al., 2011; Jönsson et al., 2006; Smith et al., 2006]) generates convergent alignments of cell polarities. The stress-based model of PIN1 polarity ([8] [Heisler et al., 2010]) also fits in the convergent alignment category as it generates similar behaviours with respect to auxin concentrations to the up-the-gradient model. The animal models that are capable of polarising in the absence of pre-established asymmetries generate tandem alignments of polarities ([4] direct coupling model [Abley et al., 2013], and (3) Burak and Shraiman, 2009).
Tables
Parameter values used in simulations of the indirect coupling model.
| Symbol | description | unit | value |
|---|---|---|---|
| Δt | numerical time step | s (seconds) | 0.05 |
| Rc | area of cytoplasmic compartment | µm2 | 260 |
| Rw | area of cell wall compartment | µm2 | 2.88 (for long cell edges); 3.0 (for short cell edges) |
| ln | length of membrane compartments | µm | 2.88; 3.0 |
| lw | length of wall compartments | µm | 2.88; 3.0 |
| cPIN | default initial concentrations of PIN1 in cytoplasmic compartments | Au/ µm2 | 0.003 |
| ρPIN | PIN1 default membrane binding rate | µm/s | 0.03 |
| τ | A*-dependent promotion of PIN1 binding | µm2/Au.s | 2 |
| µPIN | PIN1 default membrane unbinding rate | /s | 0.004 |
| DPIN | PIN1 diffusion in cell membrane | µm2/s | 0.1 |
| ΨPIN | PIN-dependent active auxin efflux rate | µm2/Au.s | 40 |
| ɛ | limit for noise addition during initialisation of A* and B* concentrations | dimensionless | 0.0166 |
| γAux | Auxin-dependent promotion of A* to A conversion | µm2/Au.s | 0.75* |
| ρAux | production rate of Auxin | Au/µm2.s | 1.0 × 10−4* |
| µAux | degradation rate of Auxin | /s | 0.01* |
| vin | influx auxin permeability | µm/s | 0.75* |
-
All parameters relate to either equations specified here or in Abley et al., 2013.
-
* In the simulations used to generate Figure 7C and Figure 7—figure supplement 1C, γAux = 0.9 µm2/Au.s, ρAux = 1.3 × 10−4 Au/µm2.s and µAux = 0.02/s. Also, in some simulations, at tissue boundaries or centres of polarity convergence, auxin production, degradation and influx auxin permeability rates may vary from the background rates. Modulation of the influx auxin permeability rate is used to simulate elevated rates of auxin import. In Figure 9C, the auxin production rate in the bottom-most row of cells = 2 × 10−4 Au/µm2.s, and the auxin degradation rate in the top most row of cells = 0.07/s. In Figure 9D, and at the beginning of the simulation used to generate Figure 10C and D the auxin production rate in the bottom-most row of cells = 2 × 10−4Au/µm2.s. Throughout both simulations, in the top-most row, the auxin degradation rate = 0.015/s and the auxin influx permeability = 4.5 µm/s. In the simulation used to generate Figure 10C and D, changes in auxin production occur during the simulation and are described below. In this simulation, in the cell that forms the centre of convergence, the inwards permeation of auxin = 37.5 µm/s and the auxin degradation rate = 0.15/s. In Figure 16C and D, the proximo-distal polarity field is established as described for Figure 9D. In the three cells with elevated auxin production, the auxin production rate is 1.4 × 10−3Au/µm2.s. In the cell with elevated auxin import and removal (Figure 16D), the auxin degradation rate = 0.1 /s and the auxin influx permeability = 37.5 µm/s. In the simulations used to generate Figure 10—figure supplement 1A and B, parameter values are as for Figures 9D and 10D, respectively except ΨPIN = 80 µm2/Au.s and γAux = 0.85 µm2/Au.s. Here, c, the concentration of A* at which half of the PINs are activated = 0.22 Au/µm and the hill coefficient, h = 2.
Parameter values used in simulations of the flux-based model.
| Symbol | description | unit | value |
|---|---|---|---|
| Δt | numerical time step | s (seconds) | 0.01 |
| R | area of cytoplasmic compartment | µm2 | 260 |
| l | length of cell edge compartment | µm | 15 for short cells edges; 8.6 for the each of the two compartments of a long edge * |
| CA | default initial concentration of auxin in cytoplasmic compartments | Au/ µm2 | 0.01 |
| CPIN | default initial concentration of PIN1 at cell edge compartments | Au /µm | 0.01† |
| ɛPIN | limit for noise addition during initialisation of PIN1 concentrations at cell edges | dimensionless | 0.025** |
| ɛAux | limit for noise addition during initialisation of Auxin concentrations | dimensionless | 0.025‡ |
| T | PIN-dependent active auxin efflux rate | µm2/Au.s | 1 |
| γ | Unbinding rate of PIN1 from the cell edge | /s | 0.1 |
| ρ | production rate of Auxin | Au/µm2.s | 0.0001§ |
| µ | degradation rate of Auxin | /s | 0.02 |
| α | Flux-dependent promotion of PIN1 allocation to a cell edge | dimensionless | 1# |
| D | Passive permeation rate of auxin | µm/s | 5 |
| Pmax | Maximum concentration of PIN1 at a cell edge | Au /µm | 0.01¶ |
| I | Auxin import rate | µm / s | 0 |
-
* For cells with hexagonal geometries, each cell edge has a length of 10 µm.
-
† Value given is that used in simulations used to generate Figure 7.B and Figure 7—figure supplement 1B. In all other simulations, CPIN = 0.
-
‡ Values apply to simulations used to generate Figure 7B (ɛPIN) and Figure 7—figure supplement 1B, and Figure 7E (ɛAux). All other simulations are initialised without noise addition.
-
§ The value given for the auxin production rate applies to simulations used to generate Figures 9A,B, 10A, B and 16A,B. In the simulation used to generate Figure 7B and Figure 7—figure supplement 1B, ρ = 0.0025 Au/µm2.s and in the simulation used to generate Figure 7E, ρ = 0.002 Au/µm2.s.
-
# The value given for, α, the flux-dependent promotion of PIN allocation to cell edges, applies to simulations used to generate Figures 9A,B, 10A,B and 16A,B. In the simulation used to generate Figure 7B, α = 4 × 10−3 and in the simulation used to generate Figure 7E, α = 3.2 × 10−3.
-
¶ In the simulation used to generate Figures 7B, Figure 7—figure supplement 1B and 7E, Pmax = 0.04Au /µm.
Parameter values used in up-the-gradient simulations.
| Symbol | description | unit | value |
|---|---|---|---|
| Δt | numerical time step | s (seconds) | 0.01 |
| R | area of cytoplasmic compartment | µm2 | 260 |
| l | length of cell edge compartment | µm | 15 for short cells edges; 8.6 for the each of the two compartments of a long edge * |
| CA | default initial concentration of auxin in cytoplasmic compartments | Au/ µm2 | 0.01 (Figure 7A,D, Figure 7—figure supplment 1A.) 0.005 (other Figs.) |
| CPIN | default initial concentration of PIN1 at cell edge compartments | Au /µm | 0.1† |
| ɛPIN | limit for noise addition during initialisation of PIN1 concentrations at cell edges | dimensionless | 0.025† |
| ɛAux | limit for noise addition during initialisation of Auxin concentrations | dimensionless | 0.025† |
| T | PIN-dependent active auxin efflux rate | µm2/Au.s | 80 |
| ρ | production rate of Auxin | Au /µm2.s | 0.0003‡ |
| µ | degradation rate of Auxin | /s | 0.005 |
| PINi | Total amount of PIN1 in a cell available for binding to edge compartments | Au /µm | 0.1 |
| D | Passive permeation rate of auxin | µm/s | 10 |
| b | Exponentiation base for PIN1 allocation to the membrane | dimensionless | 6 |
| I | Auxin import rate | µm / s | 0 |
-
* For cells with hexagonal geometries, each cell edge has a length of 10 µm.
-
† Values given for the initial concentration of PIN1 at cell edge compartments, and the limit for noise addition to this concentration, are those used in simulations used to generate Figure 7A and Figure 7—figure supplement 1A. In all other simulations, CPIN and ɛPIN = 0. The value given for ɛAux applies only to Figure 7D, in all other simulations, ɛAux = 0.
-
‡ Value given for the production rate of auxin applies to the simulations used to generate Figures 7D, 8A, B, 11H, 14D and 16E,F. In the simulation used to generate Figure 7A and Figure 7—figure supplement 1A, ρ = 0.0025 Au /µm2.s.
Additional files
-
Source code 1
Code used to generate Figure 7A.
- https://doi.org/10.7554/eLife.18165.029
-
Source code 2
Code used to generate Figure 7B.
- https://doi.org/10.7554/eLife.18165.030
-
Source code 3
Code used to generate Figure 7C.
- https://doi.org/10.7554/eLife.18165.031
-
Source code 4
Code used to generate Figure 7D.
- https://doi.org/10.7554/eLife.18165.032
-
Source code 5
Code used to generate Figure 7E.
- https://doi.org/10.7554/eLife.18165.033
-
Source code 6
Code used to generate Figure 7F.
- https://doi.org/10.7554/eLife.18165.034
-
Source code 7
Code used to generate Figure 8A.
- https://doi.org/10.7554/eLife.18165.035
-
Source code 8
Code used to generate Figure 8B.
- https://doi.org/10.7554/eLife.18165.036
-
Source code 9
Code used to generate Figure 9A.
- https://doi.org/10.7554/eLife.18165.037
-
Source code 10
Code used to generate Figure 9B.
- https://doi.org/10.7554/eLife.18165.038
-
Source code 11
Code used to generate Figure 9C.
- https://doi.org/10.7554/eLife.18165.039
-
Source code 12
Code used to generate Figure 9D.
- https://doi.org/10.7554/eLife.18165.040
-
Source code 13
Code used to generate Figure 10A and B.
- https://doi.org/10.7554/eLife.18165.041
-
Source code 14
Code used to generate Figure 10C and D.
- https://doi.org/10.7554/eLife.18165.042
-
Source code 15
Code used to generate Figure 11H.
- https://doi.org/10.7554/eLife.18165.043
-
Source code 16
Code used to generate Figure 14D.
- https://doi.org/10.7554/eLife.18165.044
-
Source code 17
Code used to generate Figure 16A.
- https://doi.org/10.7554/eLife.18165.045
-
Source code 18
Code used to generate Figure 16B.
- https://doi.org/10.7554/eLife.18165.046
-
Source code 19
Code used to generate Figure 16C.
- https://doi.org/10.7554/eLife.18165.047
-
Source code 20
Code used to generate Figure 16D.
- https://doi.org/10.7554/eLife.18165.048
-
Source code 21
Code used to generate Figure 16E.
- https://doi.org/10.7554/eLife.18165.049
-
Source code 22
Code used to generate Figure 16F.
- https://doi.org/10.7554/eLife.18165.050
-
Supplementary file 1
Supplementary model information.
Instructions on how to run models and explanation of the code for each model.
- https://doi.org/10.7554/eLife.18165.051





