Structural insights into the molecular mechanism of the m6A writer complex
Figures
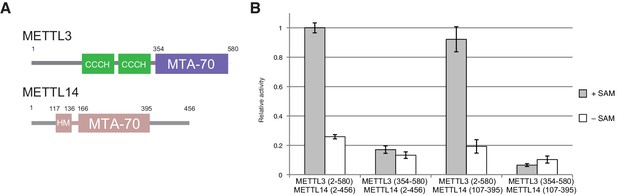
Domain composition and methyltransferase activity of METTL3-METTL14.
(A) Domain composition of METTL3 and METTL14. Predicted CCCH zinc finger motifs in the N-terminal region of METTL3 are shown in green. The crystallized METTL3-METTL14 complex contained the MTA-70 methyltransferase domain of METTL3 (residues 354–580, highlighted in blue) and a METTL14 construct (residues 117–395, highlighted in pink) comprising the MTA-70 domain preceded by an N-terminal extension (denoted HM). (B) Methyltransferase activities of the full-length METTL3-METTL14 complex, and complexes containing truncated METTL3 and METTL14 proteins used for the structure determination. The activities were determined by measuring the concentration of methylated RNA product using a m6A-specific antibody in an enzyme-linked immunosorbent assay. The activities are represented as fractions of the activity of WT complex. Reactions lacking the donor substrate SAM were performed to determine background signal in the assay due to non-specific antibody binding. Error bars represent mean ± standard error of the mean (s.e.m) of three independent replicates.
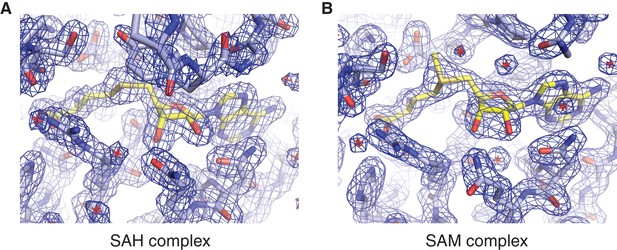
Methyl group donor substrate and product binding in the METTL3 active site.
Segments of 2Fo-Fc electron density maps corresponding to the ligand binding sites in the SAM (A) and SAH-bound (B) structures, contoured at 1.5 σ. The ligands are shown in yellow.
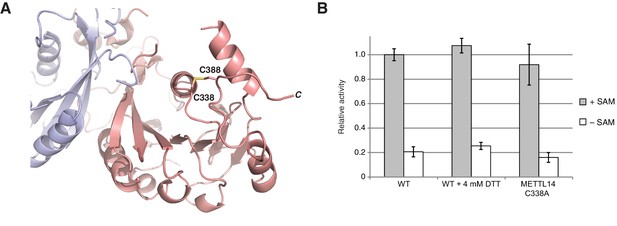
METTL3-METTL14 is catalytically active under reducing conditions.
(A) Detailed view of the disulfide bridge formed by the residues Cys338-Cys388METTL14. (B) Enzymatic activity of wild-type (WT) METTL3-METTL14 complex in the presence and absence of the reducing agent DTT and a mutant complex lacking the Cys338Mettl14-Cys388Mettl14 disulfide bridge.
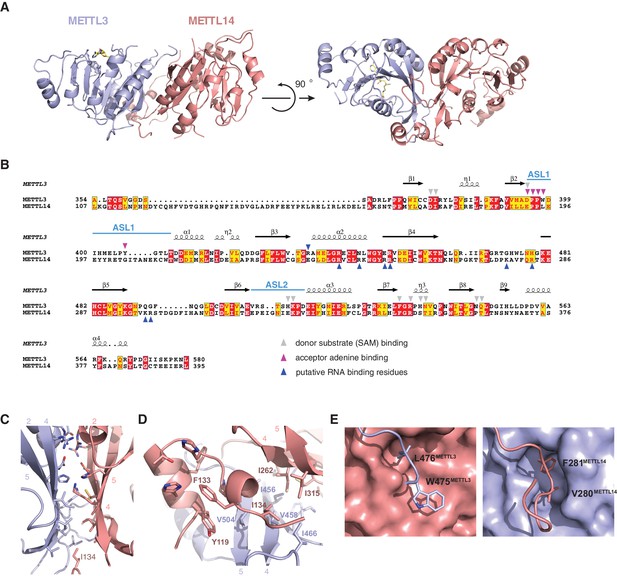
METTL3 and METTL14 form an asymmetric dimer.
(A) Overall structure of METTL3-METTL14 heterodimer. METT3 is colored blue; METTL14 is colored pink. The color coding is used throughout the manuscript. Bound SAM donor substrate is shown in stick format and colored yellow. (B) Sequence alignment of human METLL3 and METTL14. The alignment was generated using T-Coffee (Di Tommaso et al., 2011) and displayed using Espript (Gouet et al., 1999). Invariant residues are highlighted in red; conserved residues are highlighted in yellow. Amino acid residues involved in donor or acceptor substrate binding are marked with colored triangles. Secondary structure elements in METTL3 are depicted above the aligned amino acid sequences. (C) Enlarged view of the dimerization interface (orientation as in the left panel of A). (D) N-terminal helical motif of METTL14 shields the hydrophobic part of the METTL3-METTL14 interface from the solvent exposure by capping it with Ile134Mettl14. (E) Detailed views of intersubunit interactions mediated by loops Arg468-Lys480METTL3 and Lys266-Arg283METTL14 containing hydrophobic residues Trp475-Leu476METTL3 and Val280-Phe281METTL14, respectively.
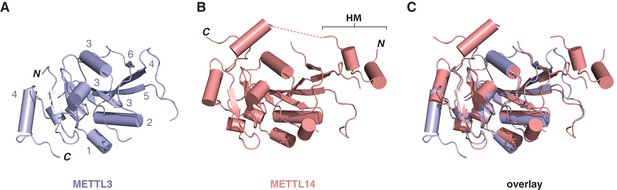
Structural comparisons of the methyltransferase domains of METTL3 and METTL14.
Structures of methyltransferase domains of METTL3 (A) and METTL14 (B), shown in the same orientation. The unstructured linked connecting the helical motif (HM) in the N-terminal extension of METTL14 with the methyltransferase domain is depicted by dashed pink line. (C) Superposition of the two methyltransferase domains.
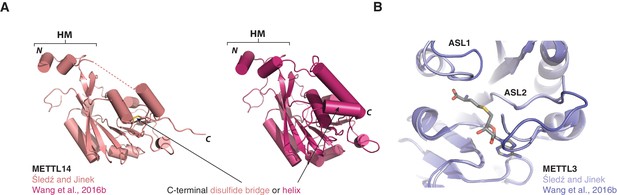
Comparisons of METTL3-METTL14 complex structures.
(A) Cartoon diagrams of METTL14 in structures determined in this study (left, light pink) and by Wang et al. (2016b) (right, colored dark purple). The structures are shown in the same orientations. The principal differences concern the presence or absence of an ordered polypeptide linker connecting the N-terminal helical motif of METTL14 (HM) and the methyltransferase domain (disordered linker indicated with a dashed line). Furthermore, due to a different C-terminal boundary, the structure reported in this study lacks a C-terminal helix in METTL14. Instead, the C-terminal region of METTL14 forms a disulfide bridge between residues Cys338METTL14 and Cys388METTL14. (B) Superposition of the METTL3 actives sites in METTL3-METTL14 structures determined in this study (left, light pink) and by Wang et al. (2016b) (right, colored dark purple).
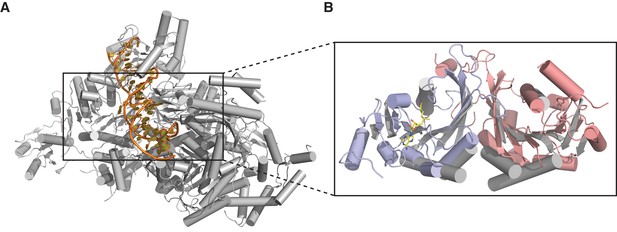
Dimerization of METTL3-METTL14 is similar to homodimeric N6-adenosine DNA methyltransferases.
(A) Structure of dimeric N6-adenosine DNA methyltransferase EcoP15I bound to a DNA substrate (Gupta et al., 2015). (B) Inset showing detail of the dimer interface of EcoP15I (colored gray) superimposed with the METTL3-METTL14 (METTL3 colored in blue, METTL14 in pink).
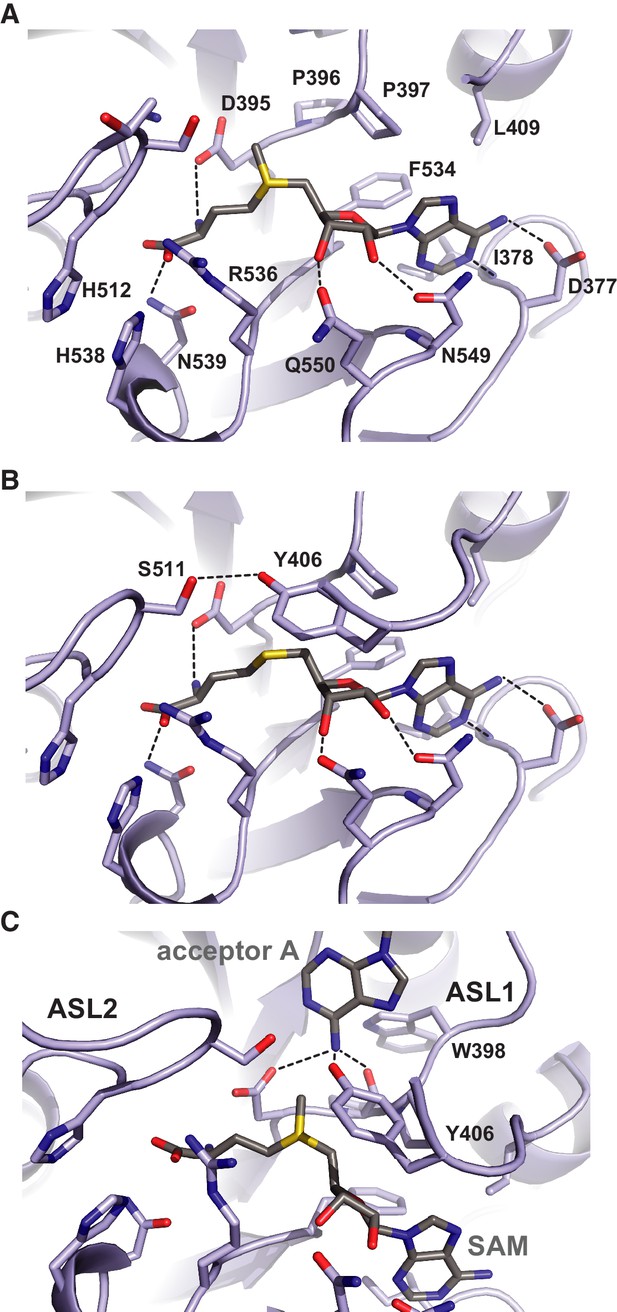
Methyl group donor substrate and product binding in the METTL3 active site.
The active site of METTL3 with (A) the donor substrate SAM and (B) product SAH bound. The ligands are shown in stick format and colored yellow. Black dashed lines indicate hydrogen bonding interactions. (C) Model of acceptor adenine binding in the METTL3 active site. Donor (SAM) and acceptor (adenine) substrates are shown in stick format and colored gray. The model was generated by superimposing METTL3 with the structure of N6-adenosine DNA methyltransferases EcoP15I (Gupta et al., 2015).
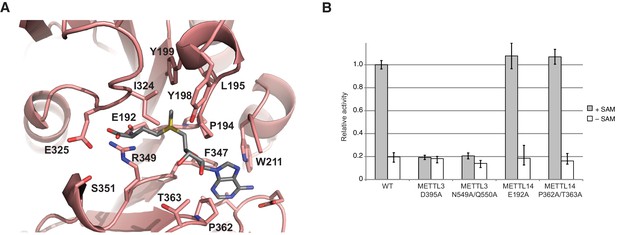
METTL14 contains a degenerate active site and is catalytically inactive.
(A) Zoom-in view of the putative methyltransferase active site in METTL14. The binding of the SAM donor cofactor (shown in grey) was modeled by superimposing the structure of SAM-bound METTL3. (B) Methyltransferase activities of the wild-type (WT) METTL3-METTL14 complex and active site alanine mutants.
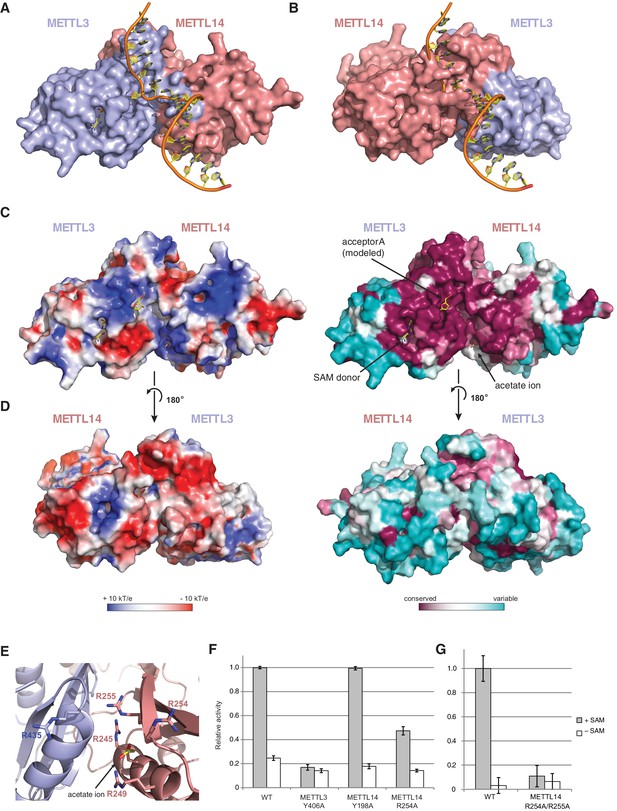
The METTL3-METTL14 dimer interface binds substrate RNA.
(A) Model of acceptor substrate binding in the active site of METTL3 obtained by superimposing METTL3 in the METTL3-METTL14 complex structure with the active methyltransferase subunit in the structure of the Mod2Res1 holocomplex of the N6-adenosine DNA methyltransferase EcoP15I bound to substrate dsDNA (PDB entry 4ZCF). For clarity, only the acceptor adenosine-containing strand of the substrate DNA is shown. METTL3-METTL14 is shown in surface representation with subunits colored as in Figure 2A. (B) Model of acceptor substrate binding to METTL14 obtained by superimposing METTL14 in the METTL3-METTL14 complex structure with the active methyltransferase subunit in the EcoP15I-dsDNA substrate complex. (C) Surface electrostatic potential (left) and sequence conservation (right) shown for the side of the METTL3-METTL14 heterodimer containing the METTL3 active site. The acetate ion and SAM cofactor co-crystallized with the protein, as well as the acceptor adenosine modeled based on EcoP15I structure are shown in yellow. The surface electrostatic potential was calculated in Pymol (Schroedinger); sequence conservation was mapped using the Consurf server (Ashkenazy et al., 2010). (D) Electrostatic surface potential and sequence conservation for the side of the complex corresponding to the degenerate METTL14 catalytic site. (E) Close-up view of the arginine-rich positively charged acetate ion binding site, proposed to interact with the ribose-phosphate backbone of substrate RNA. (F) Methyltransferase activities of WT METTL3-METTL14 complex and the alanine mutants of putative RNA substrate binding residues. The activities were measured as in Figure 1B. (G) Methyltransferase activities of WT and R254A/R255AMETTL14 mutant complex.
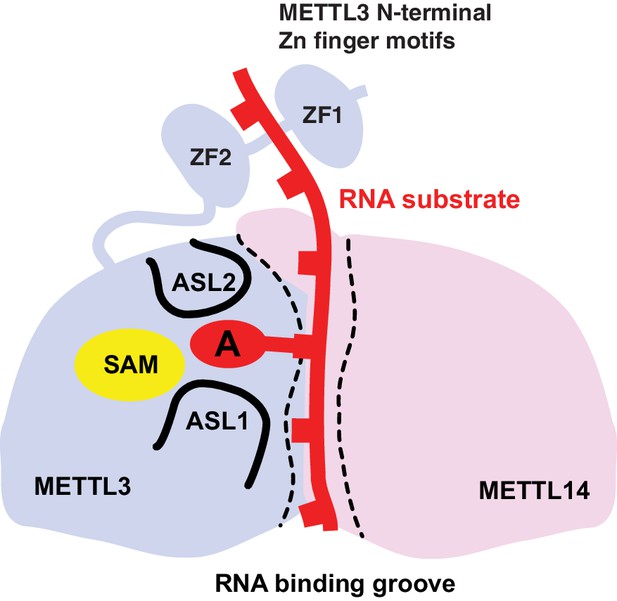
Proposed model for RNA methylation by the METTL3-METTL14 complex.
Binding of the substrate RNA to a conserved positively charged surface on the asymmetric interface of the METTL3-METTL4 heterodimer positions the acceptor adenine moiety in the catalytic pocket of METTL3 located defined by ASL1 and ASL2 loops. METTL3 catalyzes methyl group transfer from SAM to the acceptor adenine moiety. The N-terminal region in of METTL3, containing two putative CCCH zinc finger motifs, contributes to substrate RNA recognition.
Tables
Data collection and refinement statistics.
| SAM complex | SAH complex | Sulfur-SAD | |
|---|---|---|---|
| X-ray source | SLS X06DA (PXIII) | SLS X06DA (PXIII) | SLS X06DA (PXIII) |
| Space group | P 32 2 1 | P 32 2 1 | P 32 2 1 |
| Cell dimensions | |||
| a, b, c (Å) | 63.95, 63.95, 225.86 | 63.82, 63.82, 225.63 | 64.02, 64.02, 226.84 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Wavelength (Å) | 1.03965 | 1.03965 | 2.07334 |
| Resolution (Å) | (2.02–1.90) | (1.96–1.85) | (2.80–2.70) |
| Rsym (%) | 8.9 (70.6) | 7.5 (68.9) | 16.5 (97.3) |
| CC1/2 | 99.9 (89.0) | 100.0 (87.5) | 100.0 (97.9) |
| I / σI | 21.89 (3.42) | 24.51 (3.55) | 47.24 (8.03) |
| Observations | 846792 (131654) | 906619 (142053) | 3073142 (279442) |
| Unique reflections | 81250 (13087) | 87473 (14080) | 28625 (2970) |
| Multiplicity | 10.4 (10.1) | 10.4 (10.1) | 107.4 (94.1) |
| Completeness (%) | 99.9 (99.2) | 99.9 (99.3) | 100.0 (100.0) |
| Refinement | |||
| Resolution (Å) | 44.62–1.90 | 49.65–1.85 | |
| No. reflections | 43271 | 46510 | |
| Rwork/Rfree | 0.1567/0.1855 | 0.1569/0.1899 | |
| No. atoms | |||
| Protein | 3549 | 3571 | |
| Ligands | 32 | 31 | |
| Water | 359 | 405 | |
| B-factors | |||
| Mean | 28.76 | 29.89 | |
| Protein | 28.17 | 29.08 | |
| Ligands | 25.06 | 22.72 | |
| Water | 35.28 | 37.64 | |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.011 | 0.011 | |
| Bond angles (°) | 1.088 | 1.126 | |
| Ramachandran plot | |||
| % Favored | 98.1 | 98.8 | |
| % Allowed | 1.9 | 1.2 | |
| % Outliers | 0 | 0 |
-
Values in parentheses denote highest resolution shell.





