Human DNA2 possesses a cryptic DNA unwinding activity that functionally integrates with BLM or WRN helicases
Figures
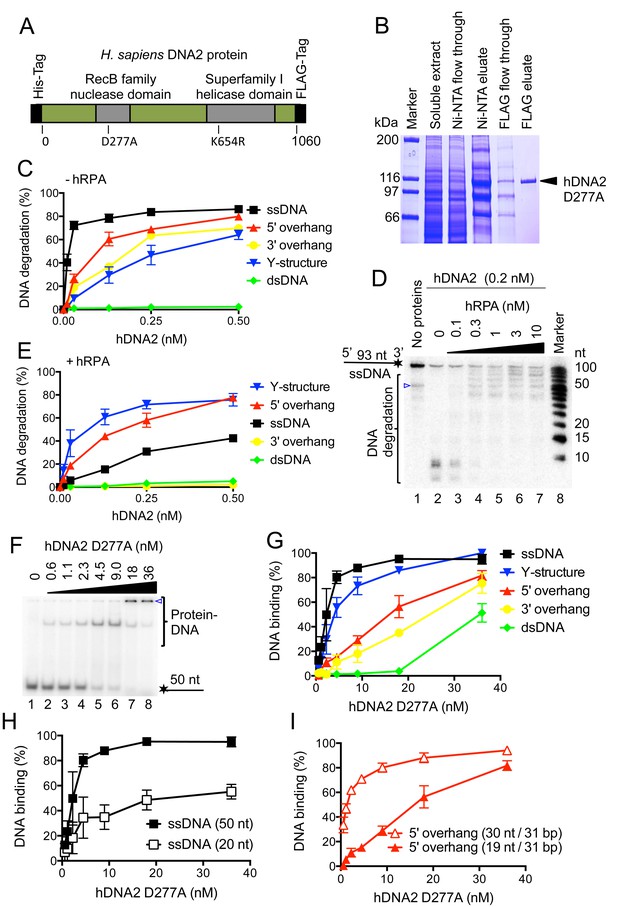
Human DNA2 preferentially binds and degrades 5’ terminated ssDNA.
(A) A schematic representation of the recombinant hDNA2 protein used in this study. The polypeptide contains an N-terminal 6xHis- and a C-terminal FLAG affinity tag. The positions of the mutations inactivating the nuclease (D277A) activity or the helicase (K654R) activity are indicated. (B) A 10% polyacrylamide gel stained with Coomassie blue showing fractions from a representative purification of hDNA2 D277A. (C) Quantitation of hDNA2 nuclease activity on various DNA substrates in the absence of hRPA from experiments such as shown in Figure 1—figure supplement 1D. Averages shown, n = 2; error bars, SEM. (D) Human DNA2 (0.2 nM) was incubated with ssDNA 32P-labeled at its 3’ end and various concentrations of hRPA. The panel shows a representative denaturing 20% polyacrylamide gel. The blue triangle indicates a truncation of the substrate. (E) Quantitation of hDNA2 nuclease activity on various DNA substrates in the presence of hRPA (15 nM) from experiments such as shown in Figure 1—figure supplement 1G. Averages shown, n = 2; error bars, SEM. (F) A representative 6% polyacrylamide gel showing the binding of hDNA2 D277A to ssDNA of 50 nt in length. The blue triangle indicates the position of the wells. (G) Quantitation of DNA binding from experiments such as shown in Figure 1F and Figure 1—figure supplement 2A–D. Averages shown, n = 2–3; error bars, SEM. (H) DNA binding and its dependence on the length of ssDNA. Quantitation is based on experiments such as shown in Figure 1F and Figure 1—figure supplement 2E. Long ssDNA was more efficiently bound by hDNA2. Averages shown, n = 2–3, error bars, SEM. (I) DNA binding and its dependence on the length of 5' single-stranded DNA overhang. Quantitation is based on experiments such as shown in Figure 1—figure supplement 2B,F. Averages shown, n = 3; error bars, SEM.
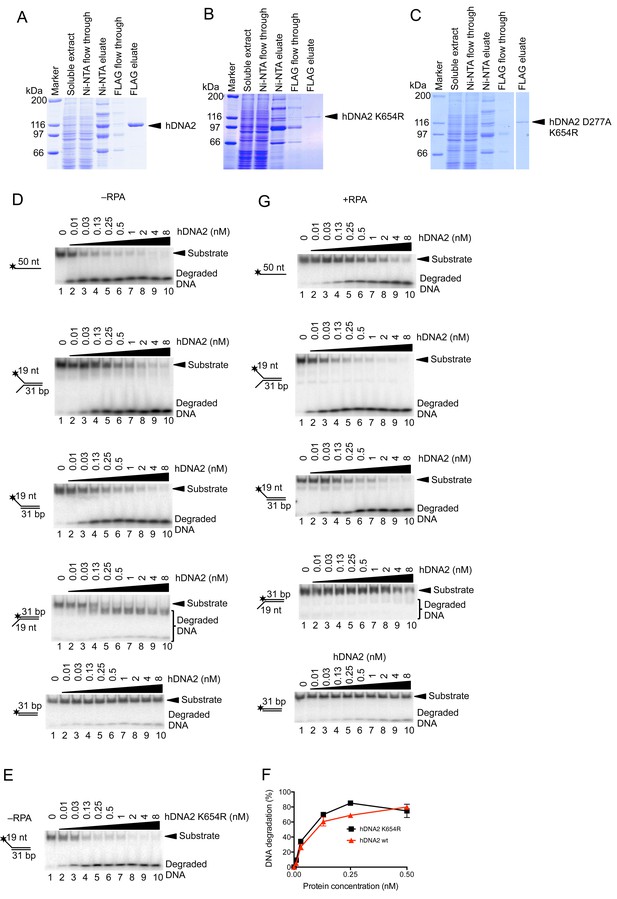
Human RPA guides the hDNA2 nuclease to 5’ terminated ssDNA.
(A–C) Representative 10% polyacrylamide gels stained with Coomassie blue showing samples from (A) wild type hDNA2, (B) helicase-deficient hDNA2 K654R, and (C) nuclease- and helicase-deficient hDNA2 D277A K654R purifications. A mutation within the helicase motif leads to reduced expression levels. (D) Representative 10% polyacrylamide gels showing the nuclease activity of wild type hDNA2 on various 32P-labeled oligonucleotide-based substrates. Reactions were carried out without hRPA. (E) Nuclease activity of the helicase-deficient hDNA2 K654R mutant. Assay as in panel D with 5' overhanged DNA substrate. (F) Quantitation of data from experiments as shown in Figure 1—figure supplement 1D,E. Averages shown, n = 2; error bars, SEM. (G) Reactions as in D but with 15 nM hRPA.
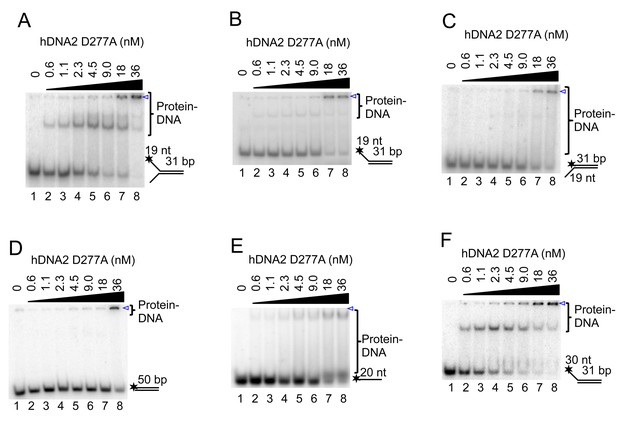
hDNA2 binds ssDNA.
(A–F) Representative 6% polyacrylamide gels showing the binding of nuclease-deficient hDNA2 D277A to various 32P-labeled oligonucleotide-based substrates. The blue triangle indicates the position of wells.
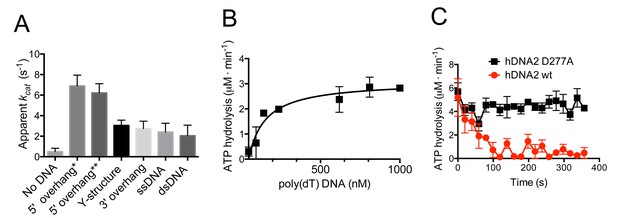
hDNA2 D277A shows DNA structure-dependent ATPase activity.
(A) Apparent ATP turnover number kcat with various DNA cofactors, including short 5’ overhang* (19 nt/31 bp), long 5’ overhang** (30 nt/31 bp), Y-structure (19 nt/ 31 bp), 3’ overhang (19 nt/ 31 bp), ssDNA (50 nt), dsDNA (31 bp). The reactions contained 12 nM hDNA2 D277A. Averages shown, n = 2–8; error bars, SEM. (B) Rate of ATP hydrolysis and its dependence on the DNA substrate concentration. The reactions contained 12 nM hDNA2 D277A and the indicated concentrations of poly(dT) DNA. Averages shown, n = 2; error bars, SEM. (C) Wild type hDNA2 or the D277A variant (both 12 nM) was incubated with 5’ overhang DNA substrate and the rate of ATP hydrolysis was determined over time. The ATP hydrolysis rate was constant at ~4–5 µM·min−1 for hDNA2 D277A and decreased over time for wild type hDNA2. Averages shown, n = 3; error bars, SEM.
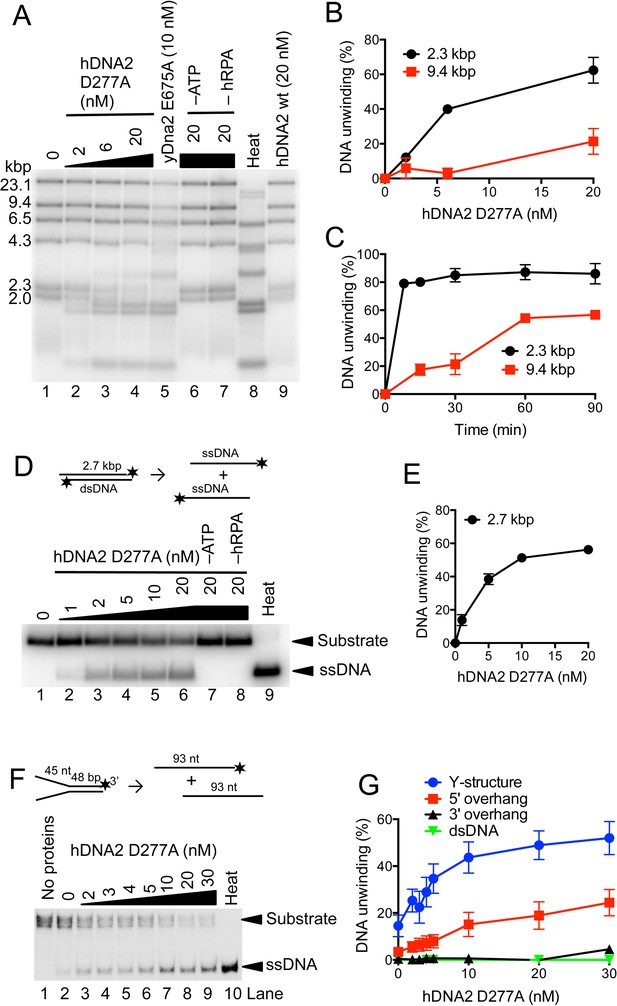
hDNA2 D277A unwinds kilobase-lengths of dsDNA.
(A) Representative 1% agarose gel showing the helicase activity of hDNA2 D277A on λDNA/HindIII substrate with 346 nM hRPA. DNA unwinding leads to products that co-migrate with heat-denatured substrate (Lane 8). Lane 5, helicase activity of nuclease-deficient yeast Dna2 E675A at 30°C; Lane 6, no ATP; Lane 7, no RPA; Lane 8, heat-denatured DNA substrate; Lane 9, wild type hDNA2. (B) Unwinding of selected λDNA/HindIII fragments by various concentrations of hDNA2 D277A upon 30 min reaction time. Quantitation of experiments such as shown in Figure 3A. Averages shown, n = 2–4; error bars, SEM. (C) Unwinding of selected λDNA/HindIII fragments by hDNA2 D277A (20 nM) and its dependence on reaction time. Quantitation of experiments such as shown in Figure 3—figure supplement 1A. Averages shown, n = 2–4; error bars, SEM. (D) Representative 1% agarose gel showing the helicase activity of hDNA2 D277A on a 2.7 kbp-long substrate. Reactions contained 215 nM RPA. Heat, heat-denatured DNA substrate. (E) Quantitation of experiments such as shown in Figure 3D. Averages shown, n = 4–9; error bars, SEM. (F) Representative 10% polyacrylamide gel showing the helicase activity of hDNA2 D277A on an oligonucleotide-based Y-structure (45 nt/ 48 bp). Reactions contained 7.5 nM RPA. Heat, heat-denatured DNA substrate. (G) Quantitation of experiments such as shown in Figure 3F and Figure 3—figure supplement 1C–E. Beside Y-structure (45 nt/48 bp), DNA substrates with 5’ or 3’ overhangs (both 45 nt/ 48 bp) and blunt-ended dsDNA (50 bp) were tested. Reactions contained 7.5 nM RPA. Averages shown, n = 2–4; error bars, SEM. Heat, heat-denatured DNA substrate.
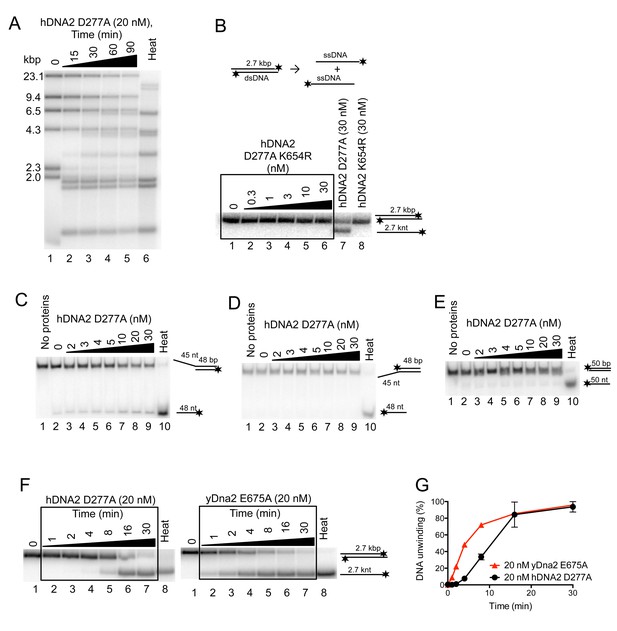
hDNA2 D277A unwinds plasmid- and oligonucleotide-based DNA substrates.
(A) Representative 1% agarose gel showing hDNA2 D277A helicase activity on a λDNA/HindIII substrate in a time-course experiment with 346 nM hRPA. Heat, heat-denatured DNA substrate. (B) Representative 1% agarose gel showing that nuclease- and helicase-deficient hDNA2 D277A K654R (lanes 2–6) and helicase-deficient hDNA2 K654R (lane 8) do not exhibit helicase activity. Lane 7, DNA unwinding by nuclease-deficient DNA2 D277A. Reactions contained 215 nM hRPA. (C–E) Representative 10% polyacrylamide gels showing the helicase activity of hDNA2 D277A with (C) 5’ overhang, (D) 3’ overhang and with (E) dsDNA substrates. Reactions contained 7.5 nM RPA. Heat, heat-denatured DNA substrate. (F) Representative 1% agarose gels showing DNA unwinding of a 2.7 kbp-long substrate by either hDNA2 D277A (left part, at 37°C) or yDna2 E675A (right part, at 30°C) in a kinetic experiment with 215 nM human RPA or 267 nM yeast RPA respectively. (G) Quantitation of experiments such as shown in F. Averages shown, n = 2; error bars, SEM.
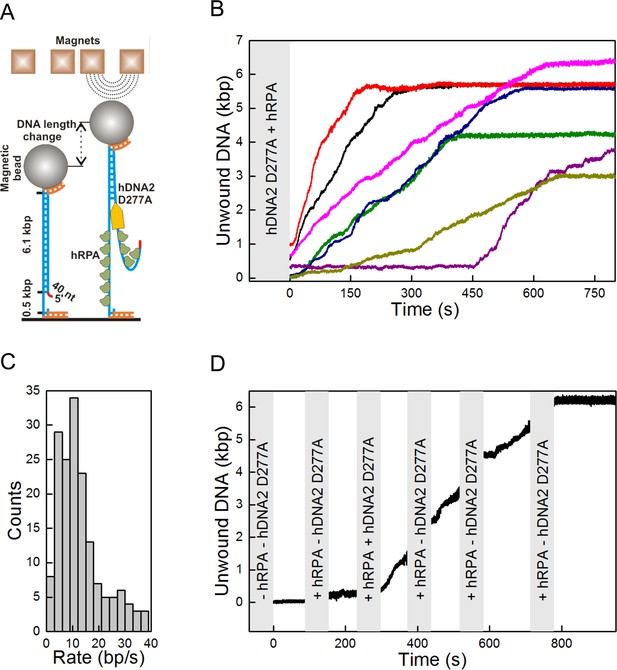
Single molecule experiments reveal highly processive DNA unwinding by hDNA2 D277A.
(A) A sketch of the magnetic tweezers assay. (B) Representative DNA unwinding events (n = 7, colored) catalyzed by hDNA2 D277A at 22 ± 3 pN force. Experiments were conducted at 37°C in a reaction buffer supplemented with 25 nM hDNA2 D277A and 25 nM hRPA. DNA lengthening was observed only after the addition of hDNA2 D277A. (C) Histogram of the observed unwinding rates. Unwinding trajectories were divided into segments with approximately constant rate. The unwinding rates of the individual segments were determined from a linear fit of the data. (D) DNA unwinding experiment at 21 pN force, initiated by adding hRPA (25 nM) at 100 s and hDNA2 D277A (25 nM) at 220 s. The buffer containing hDNA2 D277A was washed away subsequently as indicated by the gray bars.
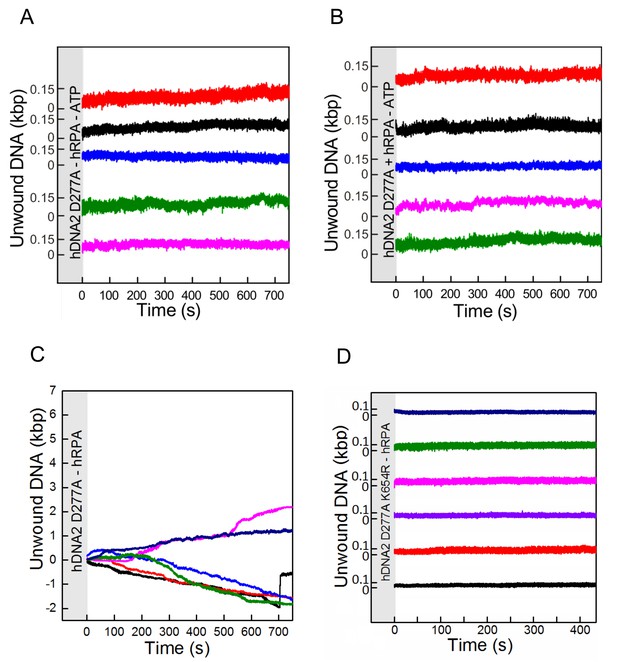
Single molecule experiments reveal that DNA unwinding by hDNA2 D277A is dependent on ATP and hRPA.
Experiments were carried out as in Figure 4B by adding hDNA2 D277A but omitting (A) both ATP and hRPA, (B) ATP or (C) hRPA only. While no activity was observed at all in the absence of ATP (independently of the presence of hRPA), some slow length changes were observed in presence of ATP but absence of hRPA that indicate a residual unwinding activity of hDNA2 D277A. This activity was dependent on the intact helicase domain of hDNA2 since experiments testing the nuclease- and helicase-deficient hDNA2 D277A K654R variant did not show such length changes as shown in (D). Length shortening may occur due to DNA looping with at least two hDNA2 molecules bound at different positions on the substrate. At this point however we have no evidence that this is physiologically relevant.
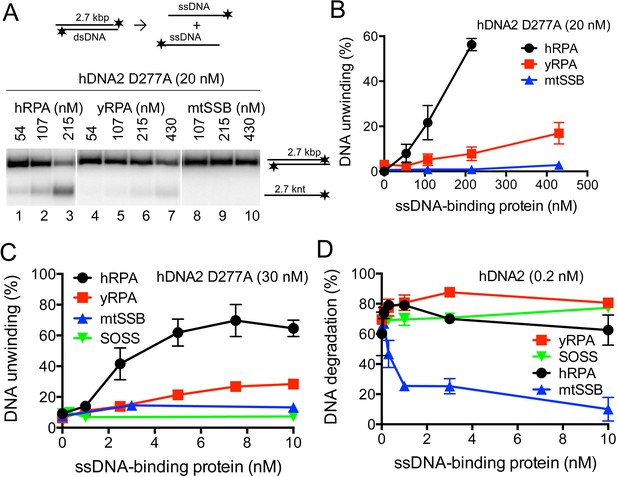
hDNA2 nuclease and helicase activities are regulated by ssDNA-binding proteins.
(A) Representative 1% agarose gels showing the helicase activity of hDNA2 D277A supplemented with indicated ssDNA-binding proteins on a 32P-labeled 2.7 kbp-long dsDNA substrate. (B) Quantitation of experiments such as shown in Figure 5A. Averages shown, n = 3–9; error bars, SEM. (C) Quantitation of unwinding experiments with Y-structured oligonucleotide-based DNA such as shown in Figure 5—figure supplement 1E. Averages shown, n = 2; error bars, SEM. (D) Quantitation of ssDNA degradation from experiments such as shown in Figure 1D and Figure 5—figure supplement 1F. Averages shown, n = 2; error bars, SEM.
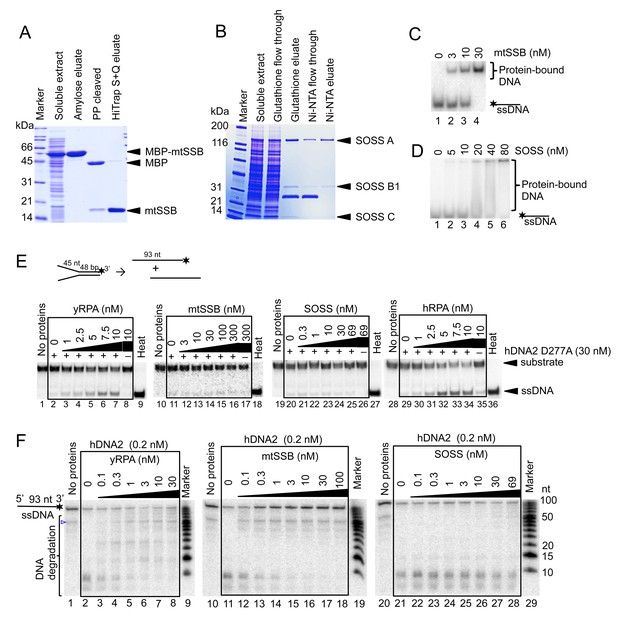
hDNA2 nuclease and helicase activities are regulated by ssDNA-binding proteins.
10% SDS-PAGE stained with Coomassie blue showing the purification procedures of (A) mtSSB and (B) the SOSS complex. PP, PreScission Protease. The ssDNA-binding properties of (C) mtSSB and (D) SOSS were tested in electrophoretic mobility shift assay. Increasing concentrations of either complex were incubated with 32P-labeled ssDNA. Representative 6% polyacrylamide gels are shown. (E) Representative 10% polyacrylamide gels showing the helicase activity of hDNA2 D277A on a 32P-labeled Y-structured DNA substrate supplemented with increasing concentrations of yRPA, mtSSB, SOSS and hRPA. 10 nM hRPA was able to melt the Y-structure to a minor degree on its own (lane 35). Heat, heat-denatured DNA substrate. Quantitation is shown in Figure 5C. (F) Representative 20% polyacrylamide denaturing urea gels showing the nuclease activity of hDNA2 (0.2 nM) on a 93 nt-long ssDNA 32P-labeled at its 3’ end. Reactions were supplemented with increasing concentrations of yRPA, mtSSB and SOSS. The blue triangle indicates a truncation of the substrate. Quantitation is shown in Figure 5D.
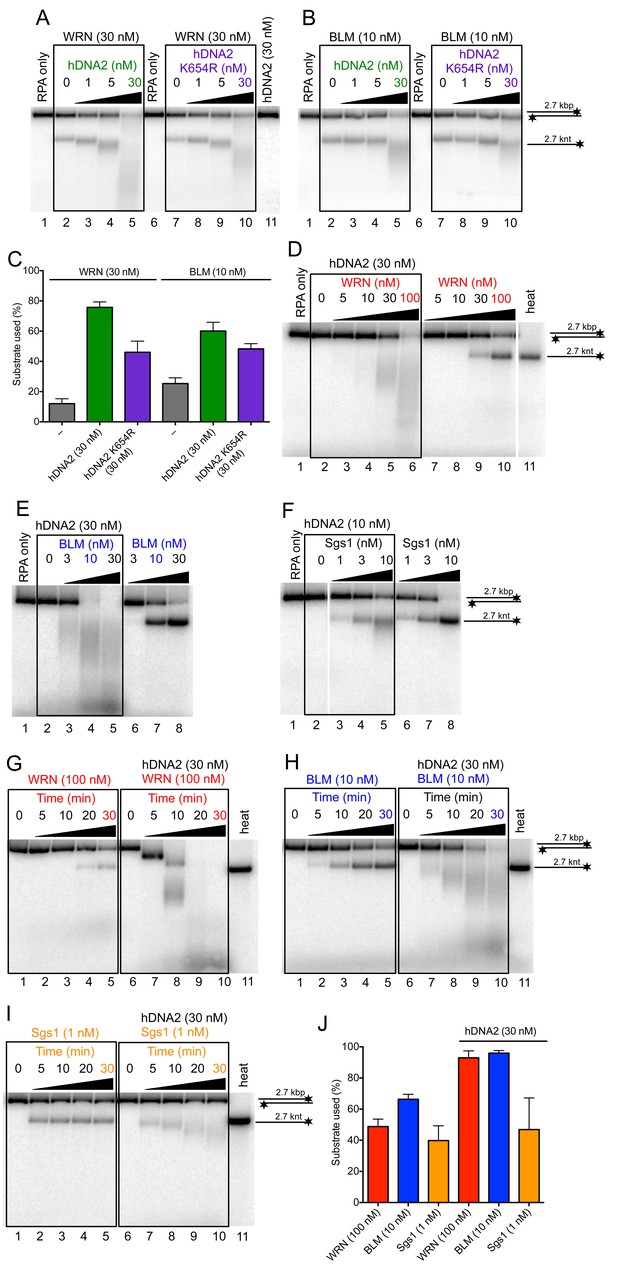
hDNA2 synergizes with WRN and BLM in the degradation of dsDNA.
Representative 1% agarose gels showing dsDNA degradation by wild type or helicase-deficient hDNA2 K654R variant with (A) WRN or (B) BLM. The reactions were supplemented with 50 mM NaCl and 215 nM hRPA. (C) Quantitation of experiments such as shown in Figure 6A,B. Averages shown, n = 4–6; error bars, SEM. Representative 1% agarose gels showing dsDNA processing by hDNA2 and (D) WRN, (E) BLM or (F) yeast Sgs1. The reactions were supplemented with 50 mM NaCl and 215 nM hRPA. Representative 1% agarose gels showing the kinetics of dsDNA processing by hDNA2 and (G) WRN (H) BLM and (I) yeast Sgs1. The reactions were supplemented with 50 mM NaCl and 215 nM hRPA. (J) Quantitation of experiments such as shown in Figure 6D,E,G–I. Averages shown, n = 3; error bars, SEM.
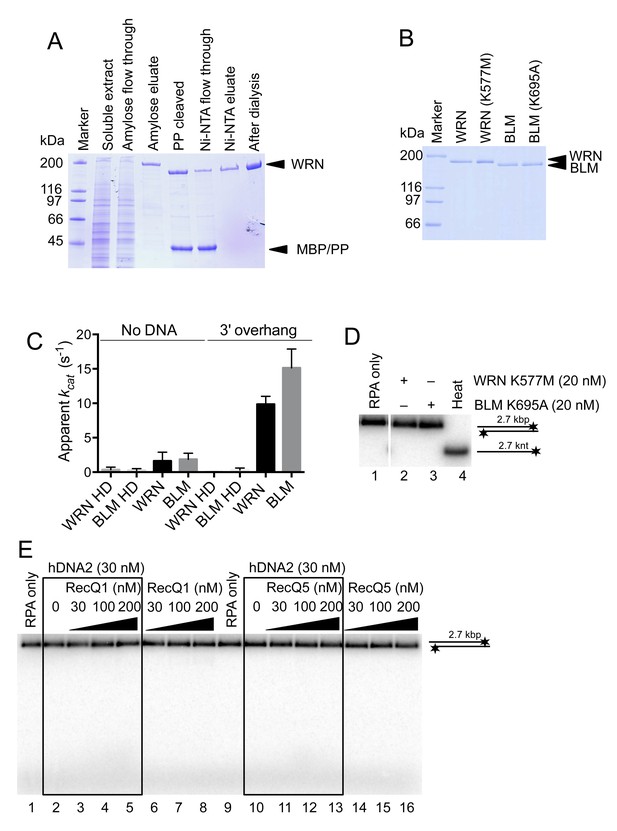
Purification of WRN and BLM proteins.
(A) 10% SDS-PAGE stained with Coomassie blue showing the purification procedure of wild type WRN. PP, PreScission Protease. (B) 10% SDS-PAGE stained with Coomassie showing wild type WRN, helicase-deficient WRN K577M, wild type BLM and helicase-deficient BLM K695A protein preparations used in this study. (C) Apparent ATP turnover number kcat showing the ATPase activity of WRN or BLM incubated with 3' overhang DNA substrate. The reactions contained 12 nM of the respective enzyme. WRN K577M and BLM K695A are devoid of ATPase activity. Averages shown, n = 2; error bars, SEM. (D) Representative 1% agarose gel showing results of unwinding assays with helicase-deficient WRN K577M and BLM K695A variants. The reactions were supplemented with 25 mM NaCl and contained 215 nM hRPA. Heat, heat-denatured DNA substrate. (E) Representative 1% agarose gel showing dsDNA degradation/unwinding reactions containing hDNA2 and RecQ1 or RecQ5 respectively. The reactions were supplemented with 50 mM NaCl and 215 nM RPA.
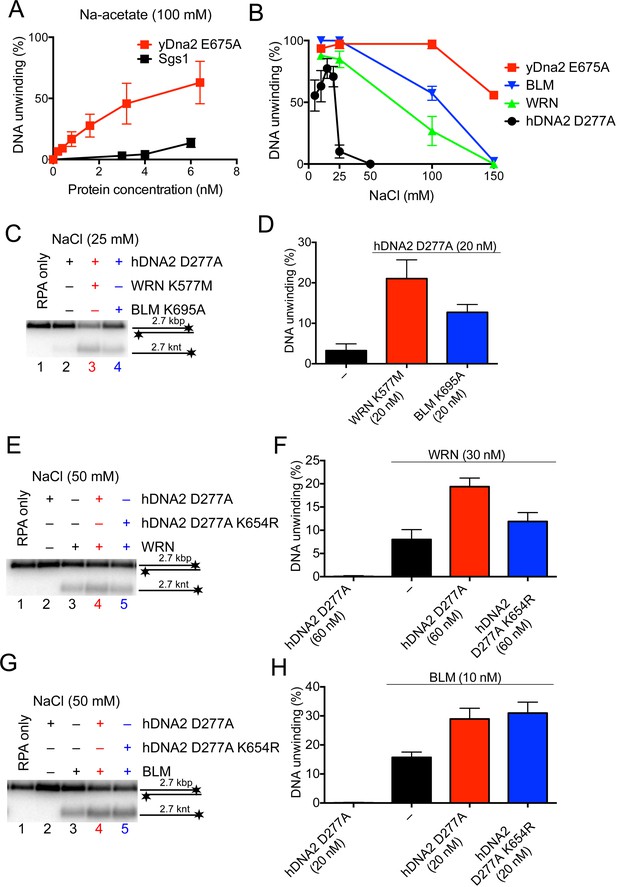
The helicase activity of hDNA2 functionally integrates with BLM or WRN helicases.
(A) Quantitation of 2.7 kbp-long dsDNA unwinding by yDna2 E675A or Sgs1 with 400 nM of yeast RPA. Reactions were supplemented with 100 mM sodium acetate and 5 mM magnesium acetate and incubated at 30°C. Averages shown, n = 2–3; error bars, SEM (B) Quantitation of DNA unwinding by yDna2 E675A (1 nM), BLM (10 nM), WRN (30 nM), hDNA2 D277A (30 nM) and its dependence on NaCl concentration. Reactions were supplemented with indicated NaCl concentrations and 2 mM magnesium acetate and incubated at 37°C. Averages shown, n = 2–3; error bars, SEM. (C) Representative 1% agarose gel showing DNA unwinding by hDNA2 D277A (20 nM) and its stimulation by helicase-deficient WRN K577M (20 nM) and BLM K695A (20 nM) variants. The reactions were supplemented with 25 mM NaCl and contained 215 nM hRPA. (D) Quantitation of experiments such as shown in Figure 7C. Averages shown, n = 5–7; error bars, SEM. (E) Representative 1% agarose gel showing the interplay of wild type WRN (30 nM) and nuclease-deficient hDNA2 D277A (60 nM) or nuclease- and helicase-deficient hDNA2 D277A K654R (60 nM) mutants. The reactions were supplemented with 50 mM NaCl and 215 nM hRPA. (F) Quantitation of experiments such as shown in Figure 7E. Averages shown, n = 3–4; error bars, SEM. (G) Representative 1% agarose gel showing the interplay of wild type BLM (10 nM) and nuclease-deficient hDNA2 D277A (20 nM) or nuclease- and helicase-deficient hDNA2 D277A K654R (20 nM) mutants. The reactions were supplemented with 50 mM NaCl and 215 nM hRPA. (H) Quantitation of experiments such as shown in Figure 7G. Averages shown, n = 2–4; error bars, SEM.
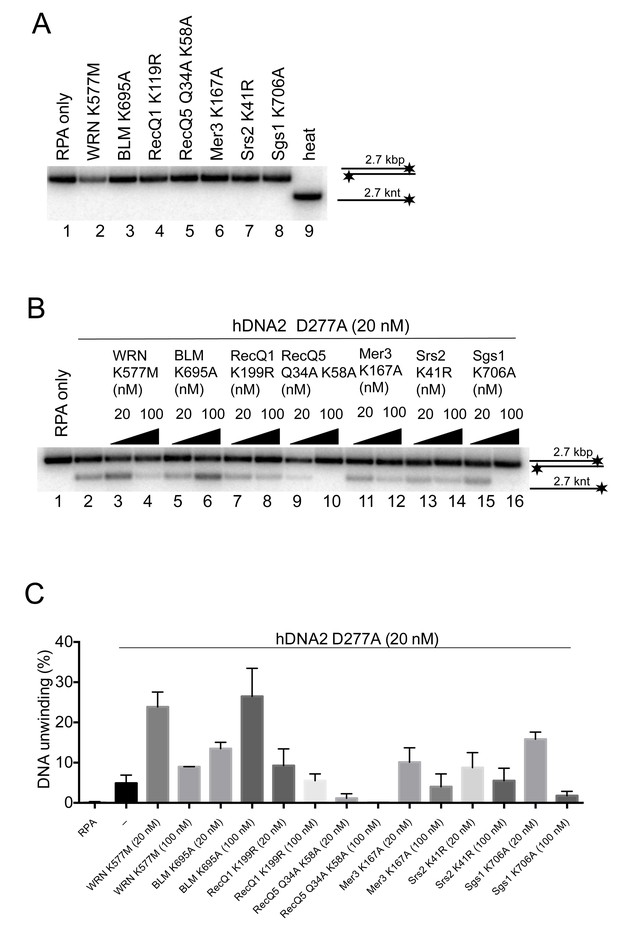
The functional integration of the helicase activity of hDNA2 is specific for WRN and BLM.
(A) Representative 1% agarose gel showing that the helicase-deficient mutants of various DNA helicases are not able to unwind a 2.7 kbp-long dsDNA substrate. (B) Representative 1% agarose gel showing DNA unwinding by hDNA2 D277A and its stimulation by helicase-deficient variants of various DNA helicases. The reactions were supplemented with 25 mM NaCl and contained 215 nM hRPA. (C) Quantitation of experiments such as shown in Figure 7—figure supplement 1. Averages shown, n = 2–8; error bars, SEM.
Additional files
-
Supplementary file 1
DNA sequences used in this study.
(A) Codon-optimized nucleotide sequence of hDNA2 gene for the expression in Sf9 cells. (B) Sequences of oligonucleotides used in this study. (C) Oligonucleotide-based DNA substrates used in this study.
- https://doi.org/10.7554/eLife.18574.016





