Transcriptional rewiring over evolutionary timescales changes quantitative and qualitative properties of gene expression
Figures
C. albicans GAL genes are needed for efficient growth on galactose.
(A) A schematic of GAL gene regulation in S. cerevisiae. (B) Log10 serial dilutions of a C. albicans parent strain (top) and isogenic strains deleted for GAL1, GAL7, and GAL10 were spotted onto plates containing 2% glucose + 3 μg/ml Antimycin A (left panel) and 2% galactose + 3 μg/ml Antimycin A (right panel). Images were acquired 6 days after growth at 30°C. The same behavior was observed for independently constructed knockout strains, as shown in Figure 1—figure supplement 1. (C) A GFP reporter strain was constructed by precisely substituting one copy of the GAL1 ORF with the GFP ORF. GFP expression of this reporter was monitored in a C. albicans parent strain, and compared with expression from isogenic strains deleted for GAL1 and orf19.6899 (a potential ortholog of S. cerevisiae GAL80). Expression was measured by flow cytometry after 6 hr of growth in the indicated media. Mean expression levels are reported for each measurement in arbitrary units. Errors indicate standard errors derived from three independent measurements.
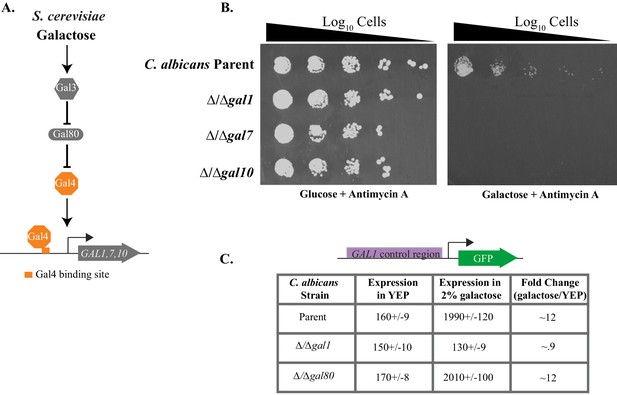
Antimycin A spotting assays in independently constructed strains of C. albicans.
(A) Log10 serial dilutions of a C. albicans reference strain (top) and knockout strains of each of the three GAL genes were spotted onto plates containing 2% galactose + 3 μg/ml Antimycin A. The strains from this image are all independently constructed from the ones shown in Figure 1B. Images were acquired 6 days after growth at 30°C. (B) Log10 serial dilutions of a C. albicans reference strain (SN250, top), knockout strains of GAL1, GAL4, CPH1, RTG1, RTG3, and addbacks of RTG1 and RTG3 were spotted onto a plate containing 2% galactose + 3 μg/ml Antimycin A. The strains from this image are all independently constructed from the ones shown in Figure 3A. The image was acquired 6 days after growth at 30°C.
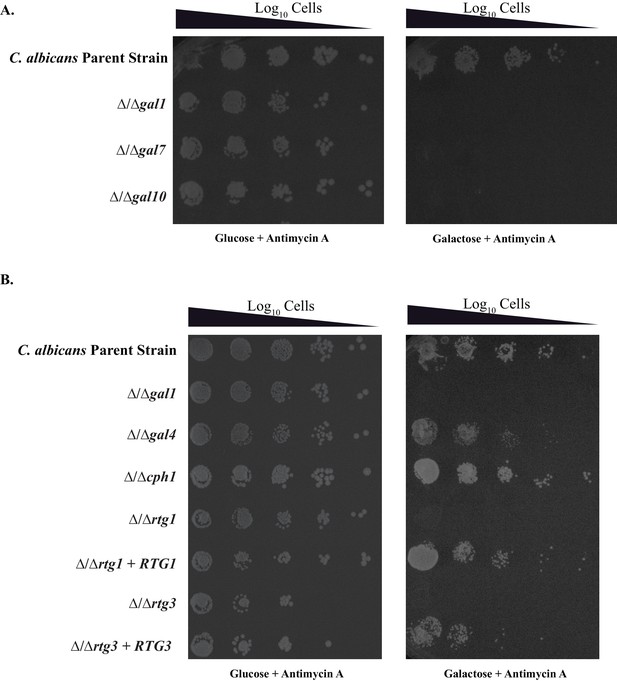
The GAL genes are required for colonization in an in vivo rat catheter model of infection.
(A–D) The parent strain (A) and the strains in which GAL1, GAL7, and GAL10 were deleted (B–D) were inoculated into rat intravenous catheters; resulting biofilms were visualized after 24 hr of growth by scanning electron microscopy. The images show the catheter luminal surfaces at 1000x magnification. The scale bar represents 20 μm.
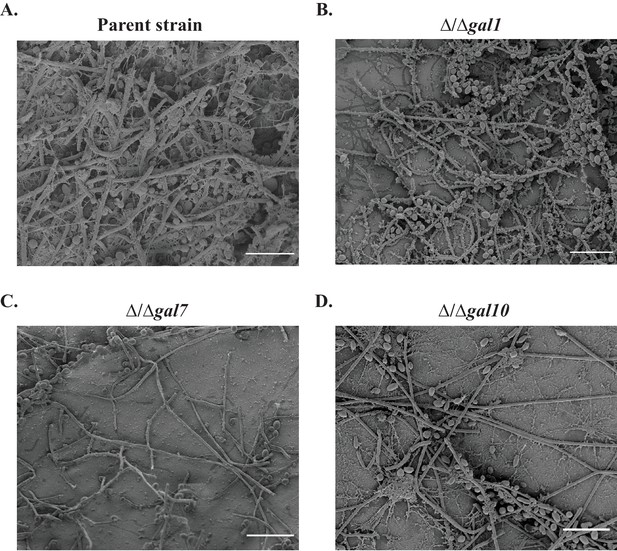
Biofilm formation in the in vivo rat catheter model.
The wild- type reference strain (A) SN425, and the (B–D) three GAL mutant strains were inoculated into rat intravenous catheters, and the resulting biofilms were visualized after 24 hr of growth by scanning electron microscopy. These images show catheter luminal surfaces at 50x magnification. The scale bar represents 100 μm.
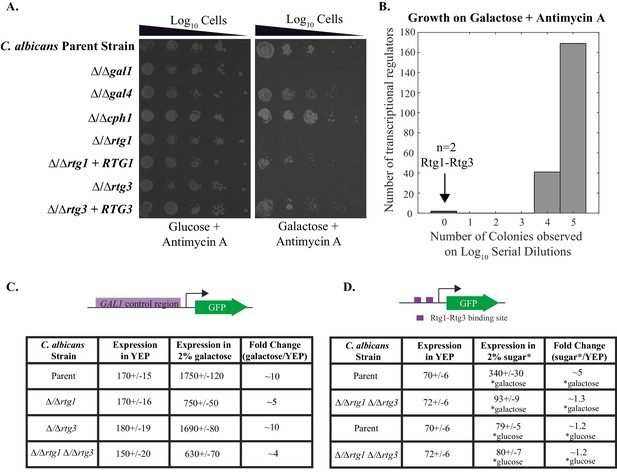
Rtg1-Rtg3 regulate galactose-mediated activation of the GAL genes in C. albicans.
(A) Log10 serial dilutions of a C. albicans parent strain (top) and isogenic strains deleted for GAL1, GAL4, CPH1, RTG1 and RTG3 were spotted onto plates containing 2% glucose + 3 μg/ml Antimycin A (left panel) and 2% galactose + 3 μg/ml Antimycin A (right panel). Also included are several gene 'addback' strains where the indicated gene was reintroduced into the corresponding deletion strain. Images were acquired 6 days after growth at 30°C. See Figure 1—figure supplement 1 for images of independently constructed isolates. (B). Log10 serial dilutions of 212 transcription factor knockout strains were spotted onto plates containing 2% galactose + 3 μg/ml Antimycin A. The numbers of dilutions where colonies were observed were tabulated and plotted as a histogram. The results show that, of the 212 deletion strains, only ∆/∆rtg1 and ∆/∆rtg3 had severe growth defects under this condition (See Figure 3—source data 1 for complete data). Neither strain showed a growth defect on plates containing 2% glucose + 3 μg/ml Antimycin A. (C) GFP expression driven by the GAL1 upstream region in a C. albicans parent strain and isogenic strains deleted for RTG1, RTG3 or both were measured by flow cytometry after 6 hr of growth in media containing 2% galactose. Mean expression levels are reported in arbitrary units with standard errors derived from three independent measurements. (D) Rtg1-Rtg3 consensus cis- regulatory motifs (found in all three of the C. albicans GAL1, GAL7 and GAL10 regulatory regions) were synthesized and ligated into a promoter lacking upstream regulatory sequences, coupled to a GFP reporter. This construct was integrated into the parent strain and isogenic strains deleted for both RTG1 and RTG3. GFP fluorescence was measured by flow cytometry after 6 hr of growth in YEP media with and without the indicated sugar. Mean expression levels are reported for each sugar in arbitrary units. Errors indicate standard errors from three independent measurements.
-
Figure 3—source data 1
Antimycin A spotting results from TFKO screen.
Colonies observed on galactose + Antimycin A and glucose + Antimycin A are tabulated for 212 strains, each one being a knockout of an individual transcriptional regulator. This data is plotted in Figure 3B.
- https://doi.org/10.7554/eLife.18981.007
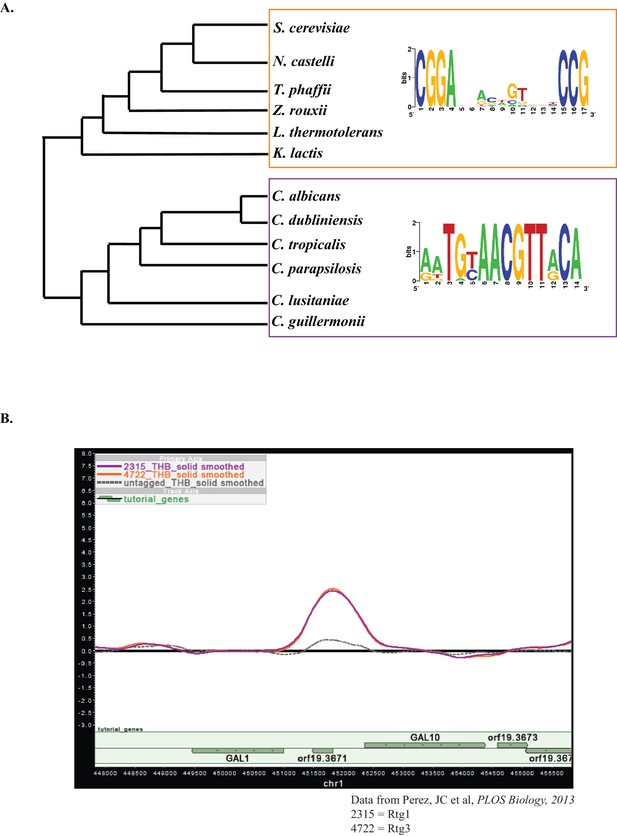
Motif analysis of GAL genes.
(A) Position-specific weight matrices for the top-scoring motifs identified by SCOPE for the GAL genes (GAL1, GAL7, GAL10) from multiple species in the CGD clade are shown (purple box). A position-specific weight matrix for the top-scoring motif identified by SCOPE for the GAL genes (GAL1, GAL7, GAL10) in S. cerevisiae and related species (orange box) are shown for comparison. We note that the phylogenetic trees are not drawn to scale. (B) Rtg1-Rtg3 binds to the GAL genes in C. albicans. A snapshot of the GAL1/GAL10 intergenic region shows binding by Rtg1 (orange line) and Rtg3 (purple line) compared to an untagged control (grey dashed line).
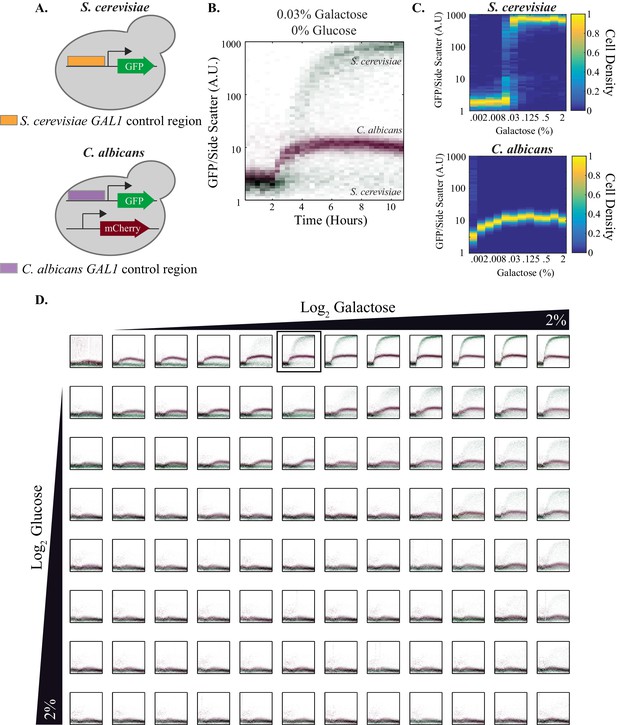
Quantitative outputs of GAL1 gene expression vary between S. cerevisiae and C. albicans.
(A) In both species, the GAL1 promoter was fused to GFP. The C. albicans strain also contained a constitutive mCherry reporter that was used to distinguish between the two species. (B) The strains were mixed together and fluorescent expression from each species was monitored for ~10 hr. The normalized fluorescence of GAL1 is plotted as a density-dependent histogram across time. Each dot (red for C. albicans and green for S. cerevisiae) represents a single cell. The panel shows a timecourse of GAL1 induction for a single concentration of galactose (and with no glucose). (C) Steady-state GAL1 gene expression of both S. cerevisiae (top) and C. albicans (bottom) is plotted as a heatmap of cell density. Cell density, the number of cells normalized by the maximum number of cells, represents the fraction of the population at a given expression level. The x-axis represents galactose concentration (in the absence of glucose) while the y- axis represents fluorescent expression. This data has been re-plotted from the panels in row 1 of Figure 4D. (D) Behavior of GAL1 across a wide range of galactose (~2000 fold) and glucose (~128 fold) concentrations, each plotted for ~10 hr. As indicated by the black wedges, galactose concentration increases left to right while glucose concentration increases from top to bottom. Red dots in each plot indicate C. albicans GAL1 expression while green dots indicate S. cerevisiae GAL1 expression. The y-axis on each plot is fluorescence, normalized by side scatter, and it spans three orders of magnitude. The data was collected every 20 min for 10 hr. The panel shown in B is indicated by the black square in the top row. See Figure 4—figure supplement 1 for additional plots from this dataset and Figure 4—figure supplement 2 for the analysis of C. albicans clinical isolates.
-
Figure 4—source data 1
Automated flow cytometry data for S. cerevisiae and C. albicans co-culture experiment across 96 galactose and/or glucose concentrations.
These are the raw flow cytometry data plotted in Figure 4D (S. cerevisiae vs. C. albicans). The file structure is setup as follows: there are several arrays for every well. Each array is a list of values obtained from each individual cell collected from that well. There is an array per flow cytometry parameter. During the course of the experiment, 32 plates of data were collected, each of which had 96 wells. Hence, the data structure contains 3072 (or 32*96) arrays.
- https://doi.org/10.7554/eLife.18981.010
-
Figure 4—source data 2
Automated flow cytometry data for C. albicans SC5314 and other C. albicans isolates experiment across galactose concentrations.
These are the raw flow cytometry data plotted in Figure 4—figure supplement 2 (C. albicans clinical isolates). The file structure is setup as follows: there are several arrays for every well. Each array is a list of values obtained from each individual cell collected from that well. There is an array per flow cytometry parameter. During the course of the experiment, 31 plates of data were collected, each of which had 96 wells. Hence, the data structure contains 2976 (or 31*96) arrays.
- https://doi.org/10.7554/eLife.18981.011
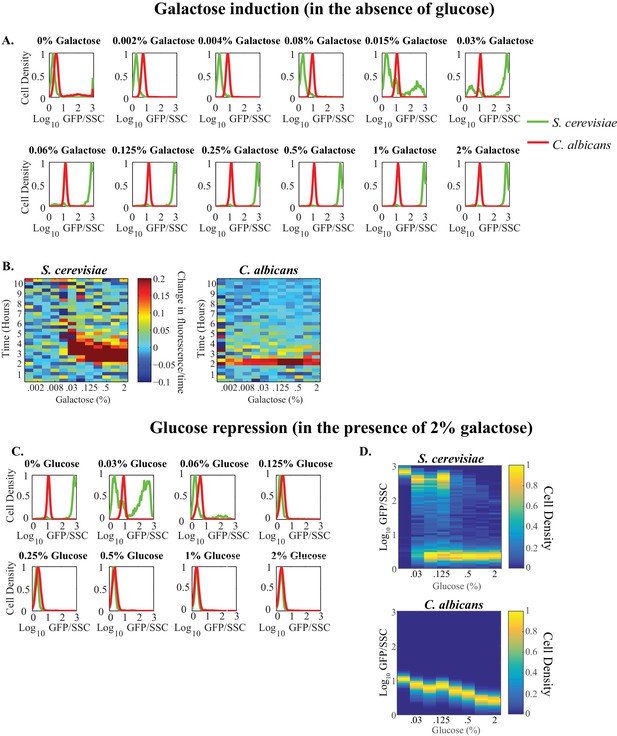
Quantitative properties of GAL gene regulation vary between S. cerevisiae and C. albicans.
(A) Steady-state histograms of GAL1 expression are plotted across a wide range of galactose (~2000 fold) concentrations in the absence of glucose. Each plot represents a specific galactose concentration. C. albicans cells are plotted in red while S. cerevisiae cells are plotted in green. The y-axis, or cell density, is the number of cells normalized by the maximum number of cells. (B) The first derivative of fluorescence for both S. cerevisiae (left) and C. albicans (right) is plotted as a heatmap to assess the timing to induction of expression. The x-axis represents galactose concentration (in the absence of glucose) while the y-axis represents time. (C) Steady-state histograms of pGAL1 expression are plotted across a wide range of glucose (~128 fold) concentrations in the presence of 2% galactose. Each plot represents a specific glucose concentration. C. albicans cells are plotted in red while S. cerevisiae cells are plotted in green. (D) Steady- state expression of both S. cerevisiae (top) and C. albicans (bottom) is plotted. The x-axis represents galactose concentration (in the presence of 2% galactose) while the y-axis represents fluorescent expression. Cell density is plotted as a heatmap.
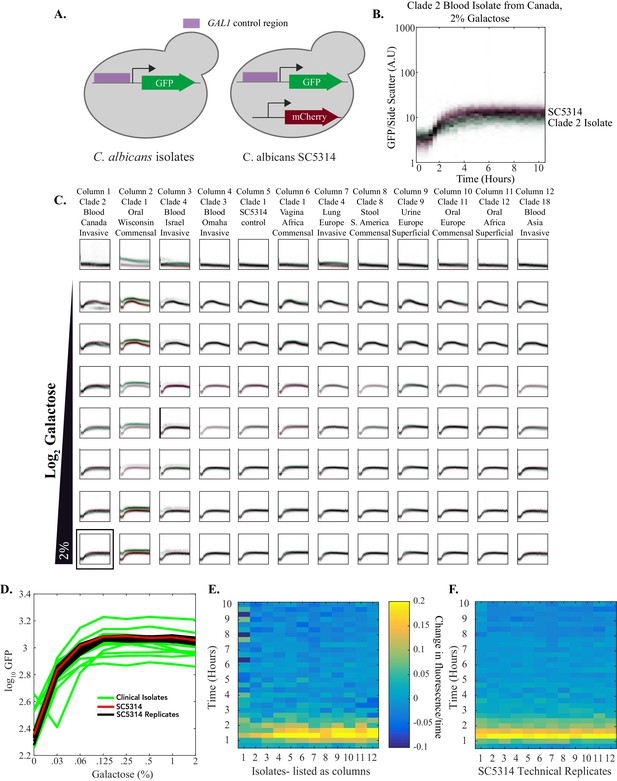
Multiple C. albicans clinical isolates show similar GAL1 gene induction in response to galactose.
(A) pGAL1 was tagged with GFP in 11 different clinical isolates (Supplementary file 1) and SC5314. These strains were competed with SC5314 containing both a pGAL1-GFP as well as a constitutively expressed mCherry. The mCherry reporter was used to distinguish the isolates from SC5314. (B). A sample plot shows how GAL1 expression is induced in response to 2% galactose. SC5314 is plotted in red while the competing clinical isolate is plotted in green. (C) GAL1 expression is plotted across ~128 fold range of galactose for each of the isolates. Each plot represents a specific galactose concentration. Each column represents a different clinical isolate while galactose concentration increases down the figure. Red dots in each plot indicate SC5314 GAL1 expression while green dots indicate GAL1 expression in a clinical isolate. The y-axis on each plot is fluorescence, normalized by size. It spans 3 orders of magnitude. The x-axis represents time. It spans 10 hr with data points every 20 min. (D) Steady-state expression of GAL1 expression is plotted as a dose-response curve. The 12 technical replicates of mCherry-tagged SC5314 are plotted in black. The non-mCherry tagged SC5314 control (column 5) is plotted in red. The 11 other clinical isolates are plotted in green. The y-axis represents a 16-fold range of fluorescent expression. It is not normalized by cell size to accentuate strain-strain variability. (E) The first derivative of fluorescence of GAL1-GFP at 0.25% galactose is plotted as a heatmap to assess the timing to induction of expression. The x-axis represents the different clinical isolates (identified by column number in Figure 4—figure supplement 2C) while the y-axis represents time. (F) The first derivative of fluorescence of GAL1-GFP at 0.25% galactose is plotted as a heatmap to assess the timing to induction of expression. The x-axis represents the different technical replicates (identified by column number in Figure 4—figure supplement 2C) while the y-axis represents time.
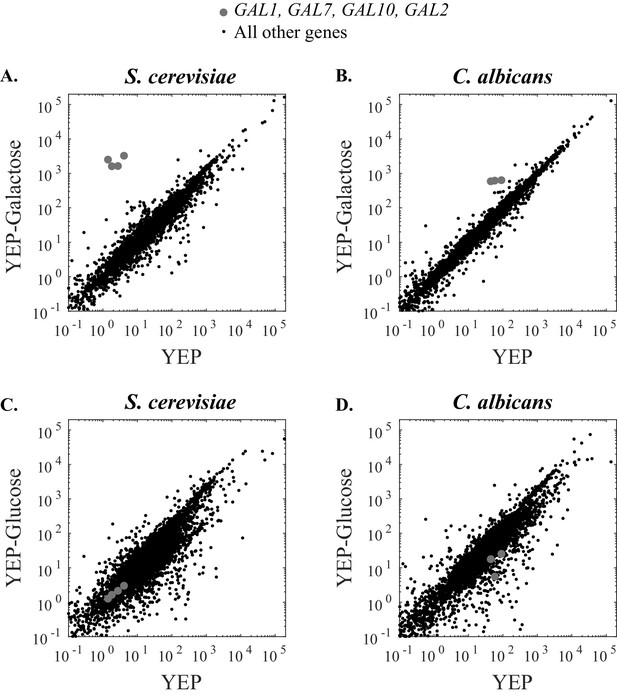
The strength of galactose induction differs between S. cerevisiae and C. albicans.
(A) RNA Sequence reads for all genes in S. cerevisiae are plotted over two conditions. The x- axis indicates the number of transcripts per million reads when cells were grown in YEP media for 6 hr while the y-axis indicates the number of transcripts per million reads when cells were grown in YEP + 2% galactose for 6 hr. The GAL1, GAL7, GAL10, and GAL2 genes are highly induced in galactose and indicated by grey dots while all other genes are shown in black. (B) RNA Sequence reads for all genes in C. albicans are plotted across the same two conditions. The GAL1, GAL7, and GAL10 genes are shown in grey (C. albicans lacks a GAL2 ortholog). (C and D) Here we show the same basic experiment, except that the comparison is between YEP and YEP + 2% glucose, rather than galactose. Additional RNA-sequencing data are provided in Figure 5—figure supplement 1.
-
Figure 5—source data 1
Quantitative analysis of S. cerevisiae RNA-Sequencing data.
Transcripts per million reads for all RNA-sequencing experiments are tabulated for all genes in three conditions. This data is plotted in Figure 5 and Figure 5—figure supplement 1.
- https://doi.org/10.7554/eLife.18981.015
-
Figure 5—source data 2
Quantitative analysis of C. albicans RNA-Sequencing data.
Transcripts per million reads for all RNA-sequencing experiments are tabulated for all genes in three conditions. This data is plotted in Figure 5 and Figure 5—figure supplement 1.
- https://doi.org/10.7554/eLife.18981.016
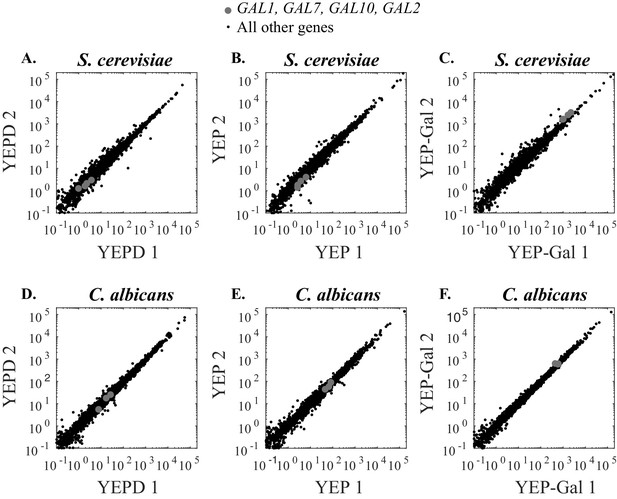
RNA-Sequencing of S. cerevisiae and C. albicans replicates in different sugar sources (glucose, no sugar, galactose).
(A–C) Scatterplots for the reads for two independent S. cerevisiae RNA-Sequencing experiments in three media conditions. GAL genes (GAL1, GAL7, GAL10, GAL2) are plotted as grey dots while all other genes are shown in black. (D–F) Scatterplots for the reads for two independent C. albicans RNA-Sequencing experiments in three media conditions. GAL genes (GAL1, GAL7, GAL10) are plotted as grey dots while all other genes are shown in black. C. albicans lacks a Gal2 ortholog.
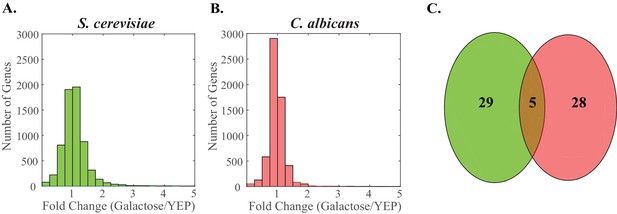
Genes induced by galactose show little overlap between S. cerevisiae and C. albicans.
In both S. cerevisiae and C. albicans, we identified the galactose regulon as the set of genes that was induced at least 2 fold (YEP + 2% galactose/YEP) in two independent RNA-Sequencing experiments. (A–B) We plot the ratio of induction for all genes in S. cerevisiae (A, green) and C. albicans (B, pink) as a histogram with a bin size of 0.25. We note that some genes induce to higher levels than the maximum of the x-axis (5) and are simply not plotted. (C) We identify overlap between these gene sets when the ortholog of a gene induced in one species is also induced in the other. Only five genes meet these criteria. This overlap is shown as a Venn diagram. The S. cerevisiae genes are shown in green (left) while the C. albicans is shown in pink (right).
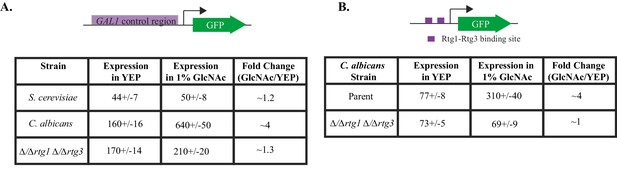
GlcNAc induces GAL gene expression in C. albicans through Rtg1-Rtg3.
(A) GAL1- GFP expression in C. albicans and S. cerevisiae was measured in cells grown in YEP +1% GlcNAc for 6 hr, compared to expression in YEP. (B) GFP expression from a C. albicans strain containing the test construct where the Rtg1-Rtg3 cis-regulatory motifs are driving a heterologous promoter was measured in similar conditions. Mean expression levels are reported in arbitrary units. Errors indicate standard errors derived from three independent measurements.
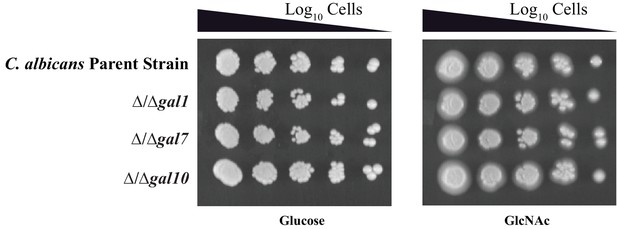
The GAL genes do not contribute to growth on GlcNAc.
Log10serial dilutions of a C. albicans parent strain (top) and isogenic strains deleted for GAL1, GAL7, and GAL10 were spotted onto plates containing 2% glucose (left panel) and 2% GlcNAc (right panel). Images were acquired 4 days after growth at 30°C.
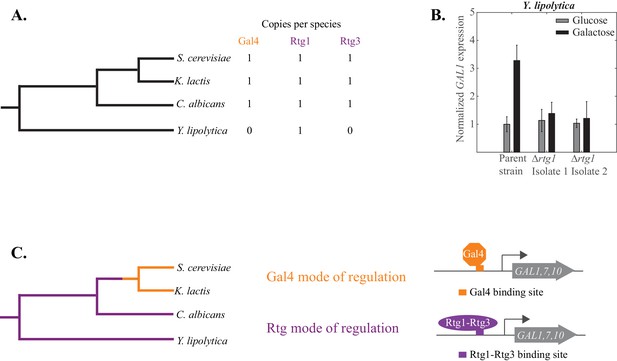
The Rtg mode of galactose-induction is ancestral to the Gal4 mode.
(A) Y. lipolytica branched before the S. cerevisiae and C. albicans clades diverged from each other. Its genome lacks an identifiable Gal4 ortholog but contains a clear Rtg1 ortholog. (B). mRNA expression of GAL1 was measured by qPCR in glucose and galactose in a Y. lipolytica parent strain and two independently constructed strains where the Rtg1 ortholog (YALI0F11979) was deleted. The relative expression level of the parent strain in glucose (compared to SGA1, a housekeeping gene) was normalized to 1 and expression of all other strains and conditions were plotted relative to it. (C) The results suggest that the Rtg mode of regulation (purple line) is ancestral to the Gal4 mode (orange line), which appeared before S. cerevisiae and K. lactis diverged from each other.
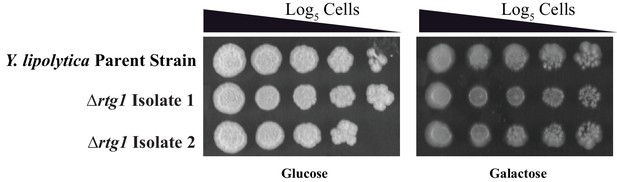
The Y. lipolytica Rtg1 ortholog does not appear to contribute to the growth of Y. lipolytica on galactose.
Log5 serial dilutions of a Y. lipolytica parent strain (top) and two independently constructed isogenic strains deleted for RTG1 (YALI0F11979) were spotted onto plates containing 2% glucose (left panel) and 2% galactose (right panel). We note that Y. lipolytica needs to respire to grow so Antimycin A cannot be used in these plating assays. Images were acquired 6 days after growth at 25°C.
Additional files
-
Supplementary file 1
Related to Figure 4—figure supplement 2: Origins of C. albicans clinical isolates.
The clades, anatomical sites of infection, geographical sites of isolation and types of infection are listed for each isolate.
- https://doi.org/10.7554/eLife.18981.023
-
Supplementary file 2
Related to Figure 5—figure supplement 2: List of genes induced by galactose at least two-fold in independent RNA-sequencing experiments in S. cerevisiae as well as gene ontology and orthology analyses of this gene set.
- https://doi.org/10.7554/eLife.18981.024
-
Supplementary file 3
Related to Figure 5—figure supplement 2: List of genes induced by galactose at least two- fold in independent RNA-sequencing experiments in C. albicans as well as gene ontology and orthology analyses of this gene set.
- https://doi.org/10.7554/eLife.18981.025
-
Supplementary file 4
C. albicans strains used in this study.
- https://doi.org/10.7554/eLife.18981.026





