Defining key roles for auxiliary proteins in an ABC transporter that maintains bacterial outer membrane lipid asymmetry
Figures
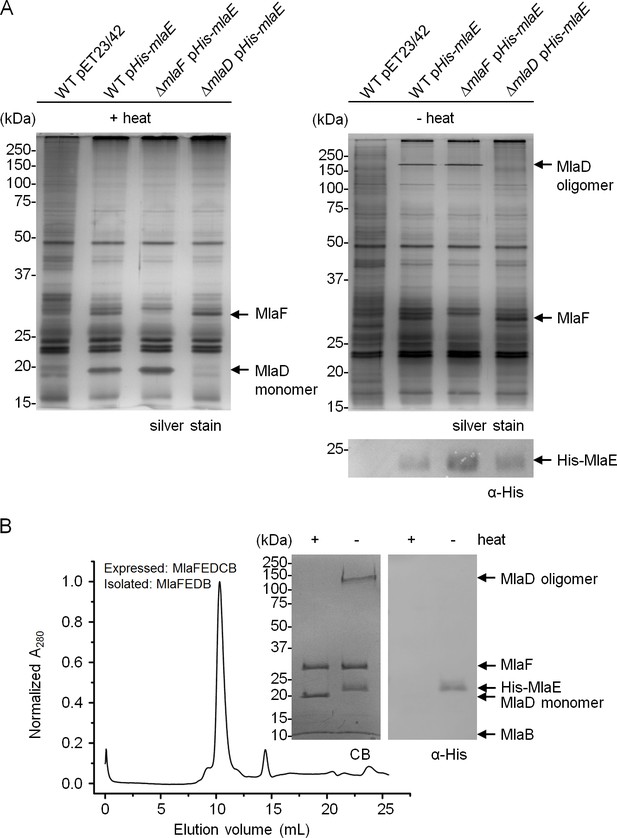
MlaF, MlaE, MlaD and MlaB form a stable complex.
(A) Co-TALON affinity purification using WT and indicated mutant strains harboring empty vector (pET23/42) or pET23/42His-mlaE (pHis-mlaE). Samples (heated or non-heated) were subjected to SDS-PAGE (12% Tris.HCl gel), and visualized by silver staining and immunoblot analyses using antibodies against the pentahistidine tag. (B) SEC profile of MlaF(His-E)DB complex purified from cells over-expressing MlaF(His-E)DCB. The peak fraction (heated or non-heated) was subjected to SDS-PAGE (4–20% Tris.HCl gel) followed by Coomassie Blue (CB) staining and immunoblot analysis. His-MlaE can only be detected on immunoblots when samples are not heated. Under the same conditions, MlaD migrates as a high molecular weight species. Positions of relevant molecular weight markers are indicated in kDa.
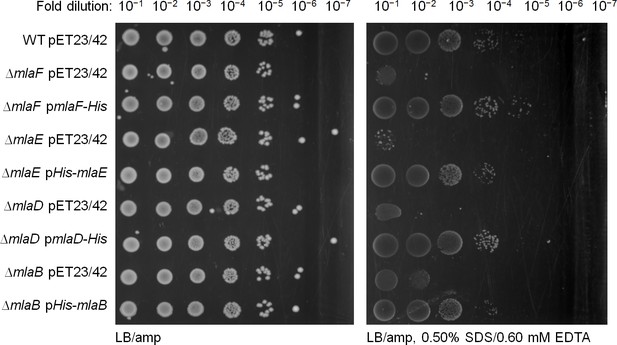
His-tagged Mla proteins are able to rescue SDS/EDTA sensitivity in the respective mla mutant strains.
Serial dilutions of cultures of wild-type (WT), ∆mlaF, ∆mlaE, ∆mlaD and ∆mlaB strains harboring pET23/42 empty vector, pET23/42mlaF-His (pmlaF-His), pET23/42His-mlaE (pHis-mlaE), pET23/42mlaD-His (pmlaD-His) or pET23/42His-mlaB (pHis-mlaB), respectively, were spotted on LB agar plates containing 200 μg/mL ampicillin, supplemented with or without 0.50% SDS and 0.60 mM EDTA as indicated, and incubated overnight at 37°C.
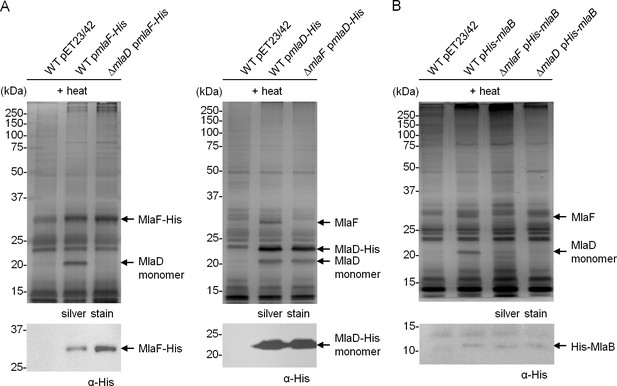
MlaF, MlaE, MlaD and MlaB form a stable complex.
Co-TALON affinity purification experiments using wild-type (WT) and indicated mutant strains harboring (A) pmlaF-His, pmlaD-His, or (B) pHis-mlaB. Samples were heated and subjected to SDS-PAGE (12% Tris.HCl gel), and visualized by silver staining and immunoblot analyses using antibodies against the pentahistidine tag. Positions of relevant molecular weight markers are indicated in kDa.
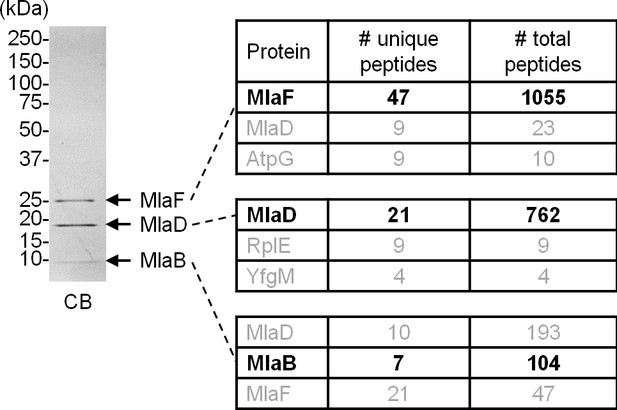
MlaF, MlaD and MlaB co-purify with His-tagged MlaE following overexpression and affinity purification.
Tandem MS/MS sequencing confirmed the identities of protein bands corresponding to MlaF, MlaD and MlaB in a preparation of the purified MlaFEDB complex. The top three most abundant proteins in each band are shown. Positions of relevant molecular weight markers are indicated in kDa.
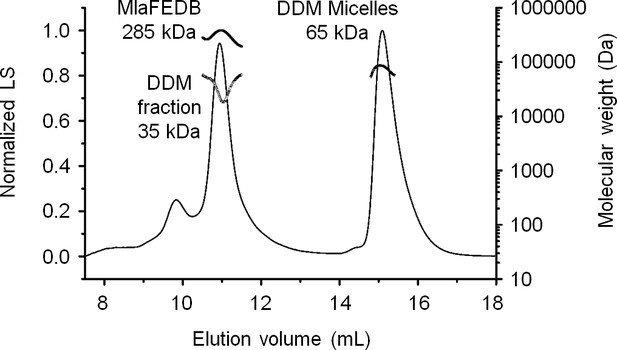
SEC-MALS analysis of the MlaF(His-E)DB complex.
Molecular mass: 256 kDa (predicted, MlaF2E2D6B2), 285 ( ± 0.5%) kDa (observed). Molecular masses of DDM fraction in complex and free micelles are 35 ( ± 4.4%) kDa and 65 ( ± 2.1%) kDa, respectively. Numbers stated after ± show statistical consistency of analysis.
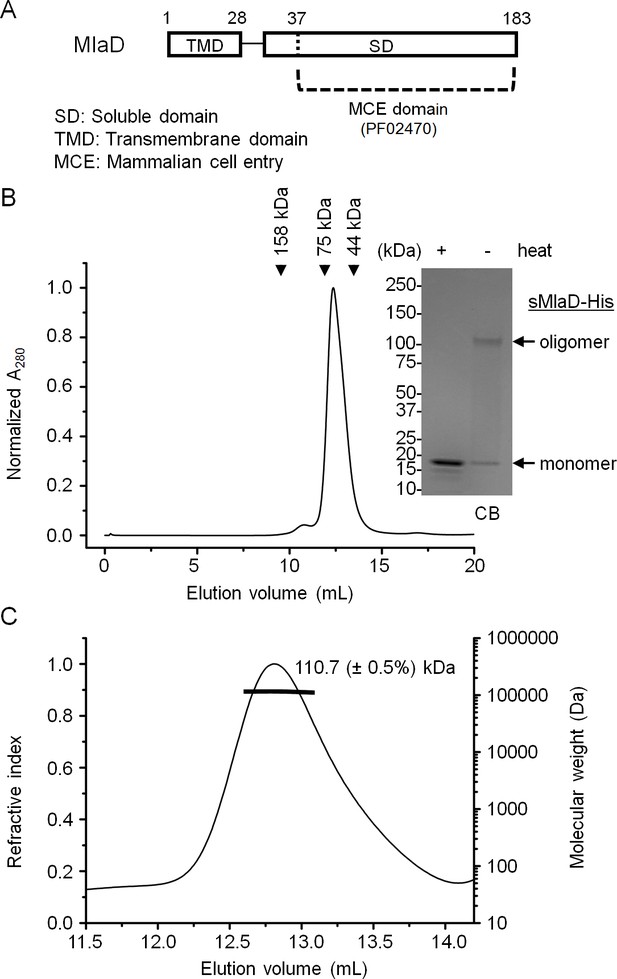
MlaD forms SDS-resistant hexamers via its soluble domain.
(A) Domain organization of MlaD. (B) SEC profile of purified soluble domain of MlaD (sMlaD-His). Elution volumes of standard globular proteins (aldolase 158 kDa, conalbumin 75 kDa and ovalbumin 44 kDa) are indicated. The peak fraction (heated or non-heated) was subjected to SDS-PAGE (4–20% Tris.HCl gel) followed by CB staining. Positions of relevant molecular weight markers are indicated in kDa. (C) SEC-MALS analysis of sMlaD-His. Hexamer molecular mass: 107 kDa (predicted), 110.7 ( ± 0.5%) kDa (observed). Numbers stated after ± show statistical consistency of the analysis.
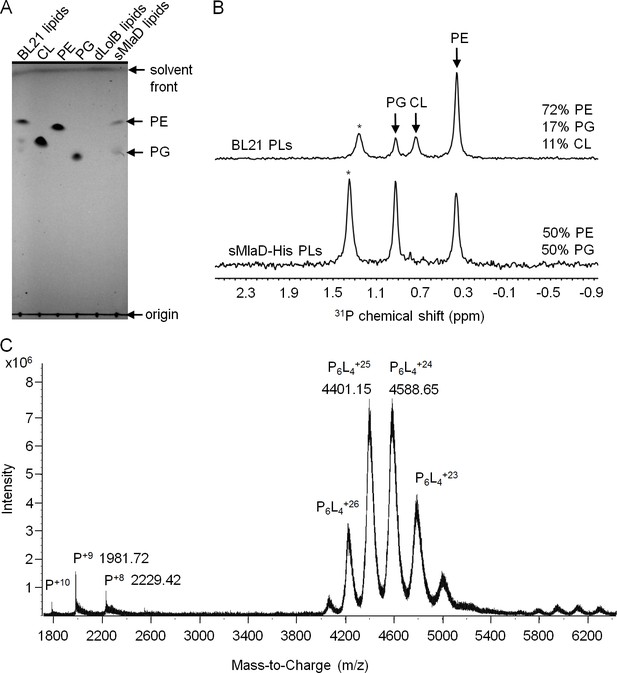
sMlaD co-purifies with endogenous PLs.
(A) TLC analysis of PLs extracted from BL21(λDE3) cells, purified dLolB-His and sMlaD-His. (B) 31P NMR analysis of PL extracts from BL21(λDE3) cells and purified sMlaD-His in 5% Triton X-100. Compositions of bound PLs were obtained via integration of peak areas, and normalized to the number of phosphorus atoms per PL molecule (i.e. one for PE/PG and two for CL). Unknown peaks that cannot be assigned to any PL species in E. coli (see Figure 3—figure supplement 1) are annotated with asterisks (*). (C) Positive mode, non-denaturing electrospray ionization (ESI) mass spectrum of sMlaD-His. Under native conditions (20 mM ammonium acetate, pH 6.9), sMlaD hexamers with charge states centred around +24 could be detected. After deconvolution, the molecular weight of the native hexamers was ~110 kDa, indicating the presence of at least four bound PLs (i.e. P6L4,assuming an average mass of 750 Da per PL).
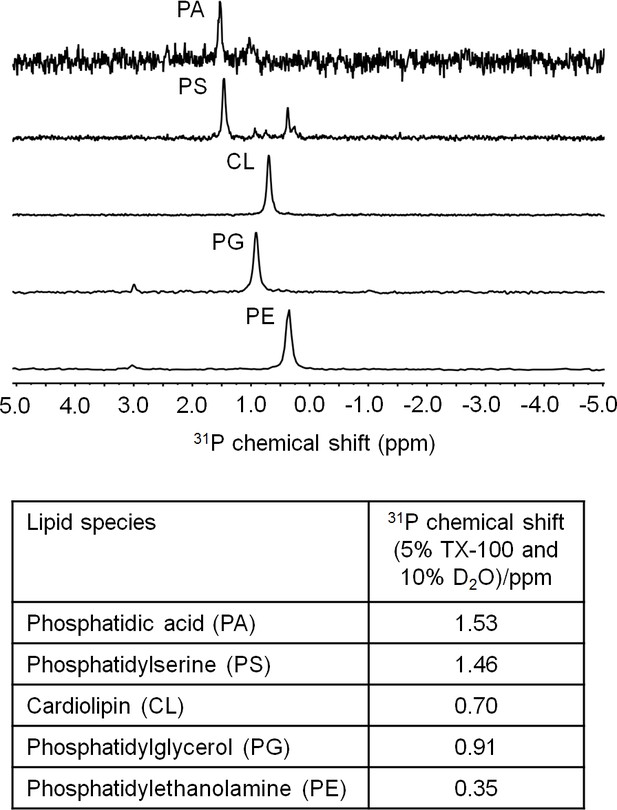
31P NMR analysis of E. coli PLs. 31P chemical shifts (ppm) of phosphatidic acid (14:0) (PA), phosphatidylserine (PS), cardiolipin (CL), phosphatidylglycerol (PG), and phosphatidylethanolamine (PE) are given in the table.
PS were extracted from E. coli EH150 strain, which accumulates PS when grown at 42°C (Hawrot et al., 1975).
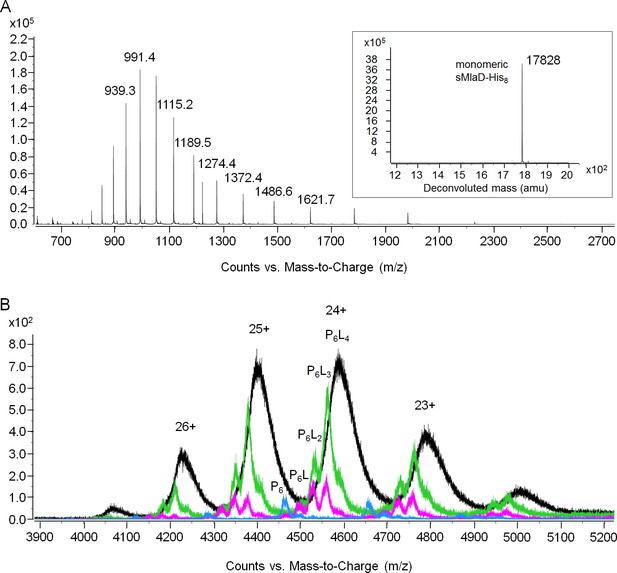
MS analyses of sMlaD-His.
(A) Positive-mode ESI-MS spectrum of sMlaD-His under denaturing conditions (50% acetonitrile 0.2% formic acid), revealing monomeric sMlaD-His at various charge states. Inset: Deconvoluted spectrum of monomeric sMlaD-His(experimental 17828.32 Da, theoretical 17827.77 Da). (B) Positive mode ESI-MS spectrum of sMlaD-His under non-denaturing conditions (20 mM ammonium acetate, pH 6.9) with increasing collision energies (0 V (black), 80 V (green), 100 V (pink) and 150 V (blue)), progressively revealing hexameric sMlaD-His with four to zero bound PL molecules.
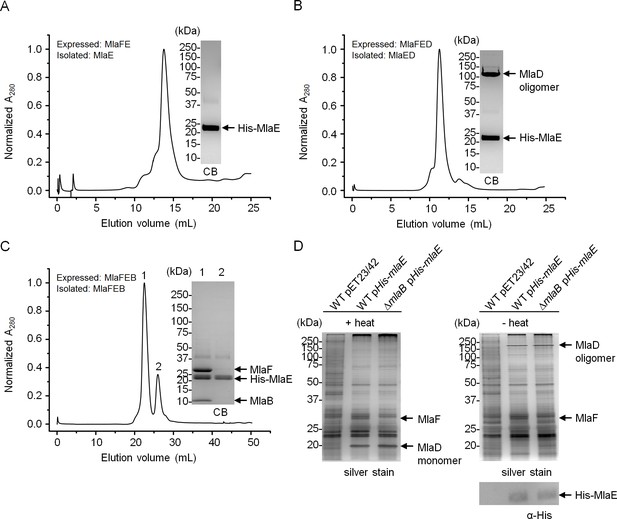
MlaB is required for the stability and/or assembly of the canonical ABC transporter.
SEC profiles of (A) His-MlaE purified from cells over-expressing MlaF(His-E), (B) (His-MlaE)D purified from cells over-expressing MlaF(His-E)D, and (C) MlaF(His-MlaE)B purified from cells over-expressing MlaF(His-E)B. The respective peak fractions (non-heated) were subjected to SDS-PAGE (4–20% Tris.HCl gel) followed by CB staining. (D) Co-TALON affinity purification using WT and ∆mlaB strains harboring empty vector (pET23/42) or pET23/42His-mlaE (pHis-mlaE). Samples (heated or non-heated) were subjected to SDS-PAGE (12% Tris.HCl gel), and visualized by silver staining and immunoblot analyses using antibodies against the pentahistidine tag. Positions of relevant molecular weight markers are indicated in kDa.
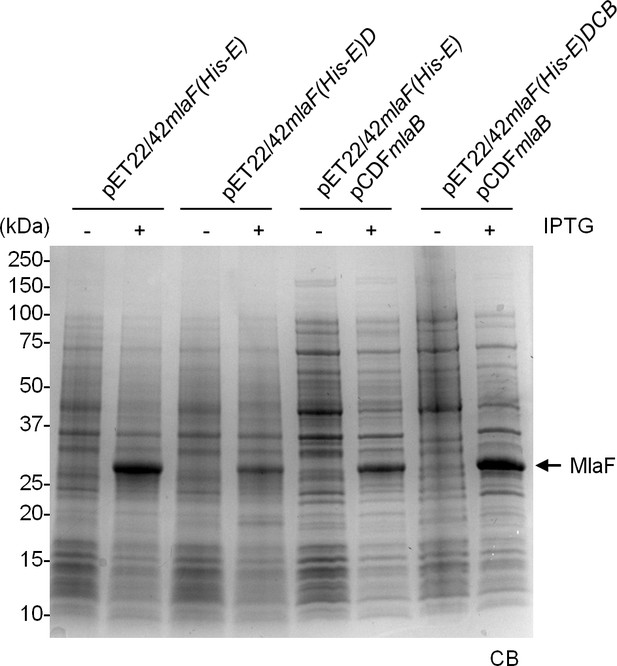
MlaF is produced at high levels in strains over-expressing full and sub-complexes of the IM ABC transporter.
SDS-PAGE (12% Tris.HCl gel) analysis of cell lysates isolated from strains over-expressing MlaF(His-E), MlaF(His-E)D, MlaF(His-E)B and MlaF(His-E)DCB from indicated vectors, with or without IPTG induction. Positions of relevant molecular weight markers are indicated in kDa.
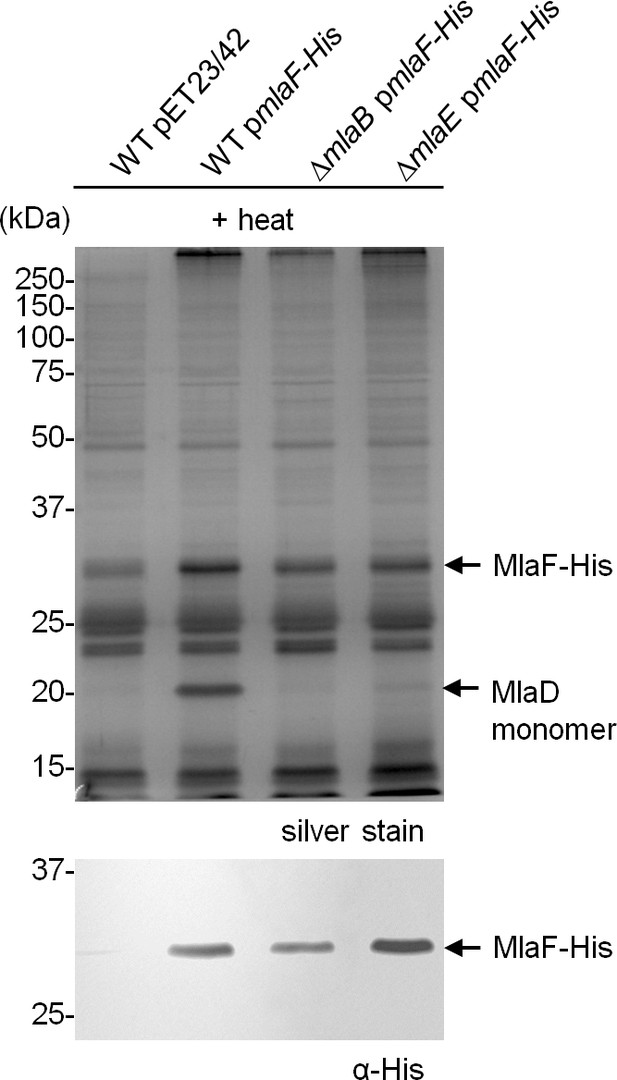
MlaD is not co-purified with MlaF-His in the absence of MlaB.
Co-TALON affinity purification using wild-type (WT), ∆mlaB and ∆mlaE strains harboring an empty pET23/42 vector or pmlaF-His. Samples were heated and subjected to SDS-PAGE (12% Tris.HCl gel), and visualized by silver staining and immunoblot analyses using antibodies against the pentahistidine tag. Positions of relevant molecular weight markers are indicated in kDa.
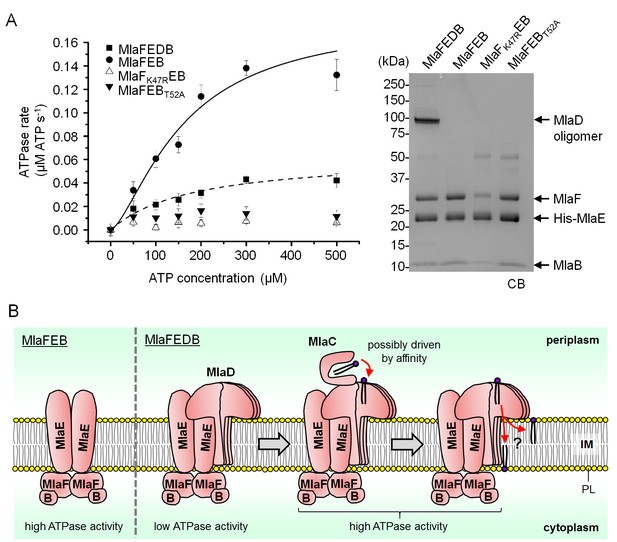
MlaD and MlaB modulate ATP hydrolytic activity of the IM ABC transporter.
(A) Enzyme-coupled ATPase assays of indicated complexes (0.1 μM) performed in detergent micelles (0.05% DDM). Average ATP hydrolysis rates (obtained from triplicate experiments, see Figure 5—source data 1) were plotted against ATP concentrations, and fitted to an expanded Michaelis-Menten equation that includes a term for Hill coefficient (n); MlaFEDB (kcat = Vmax/[complex] = 0.6 ± 0.3 μmol ATP s-1/μmol complex, Km = 181.1 ± 203.6 μM, n = 1.0 ± 0.6) and MlaFEB (kcat = 1.8 ± 0.5 μmol ATP s-1/μmol complex, Km = 161.4 ± 75.57 μM, n = 1.5 ± 0.5). SDS-PAGE analysis of the complexes (non-heated) used for these assays is shown on the right. Error bars in the graph and numbers stated after ± are standard deviations of triplicate data. (B) A proposed model for how the MlaFEDB complex functions to drive PL transport from the OM to the IM.
-
Figure 5—source data 1
Source data for ATPase assay.
- https://doi.org/10.7554/eLife.19042.016
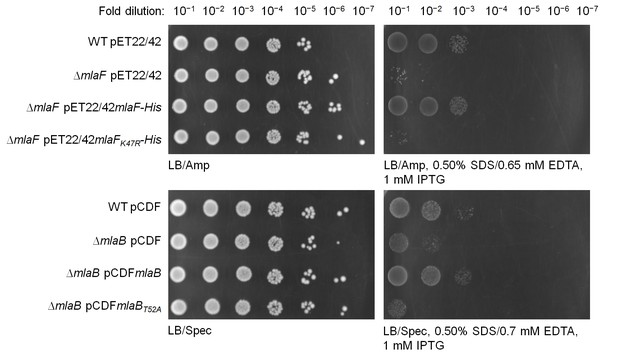
mlaFK47R and mlaBT52A are non-functional alleles.
Serial dilutions of cultures of wild-type (WT) and ∆mlaF strains harboring pET22/42 empty vector, pET22/42mlaF-His or pET22/42mlaFK47R-His, or WT and ∆mlaB strains harboring pCDF empty vector, pCDFmlaB or pCDFmlaBT52A were spotted on LB agar plates containing appropriate antibiotics, supplemented with or without 0.50% SDS, 0.65/0.70 mM EDTA as indicated, and incubated overnight at 37°C. The strains used here do not express T7 polymerase. IPTG (1 mM) was added to allow de-repression and thus low-level transcription (by endogenous polymerases) from the T7-lacO promoter in these plasmids.
Additional files
-
Supplementary file 1
Bacteria strains used in this study.
- https://doi.org/10.7554/eLife.19042.018
-
Supplementary file 2
Plasmids used in this study.
- https://doi.org/10.7554/eLife.19042.019
-
Supplementary file 3
Primers used in this study.
- https://doi.org/10.7554/eLife.19042.020





