Factors essential for L,D-transpeptidase-mediated peptidoglycan cross-linking and β-lactam resistance in Escherichia coli
Figures
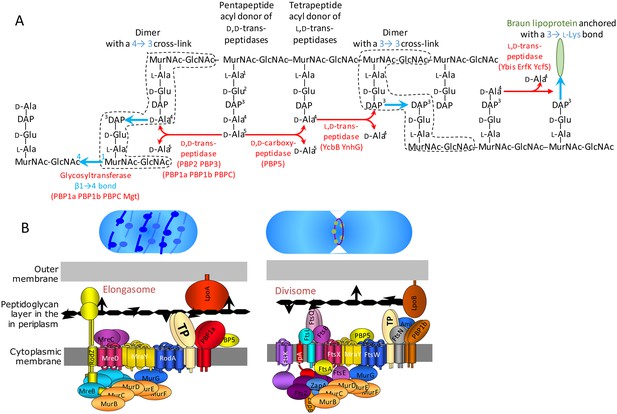
Peptidoglycan synthesis in E. coli.
(A) Reactions catalyzed by enzymes involved in peptidoglycan polymerization and Braun lipoprotein anchoring. (B) Complexes responsible for peptidoglycan synthesis during lateral cell-wall growth and division.
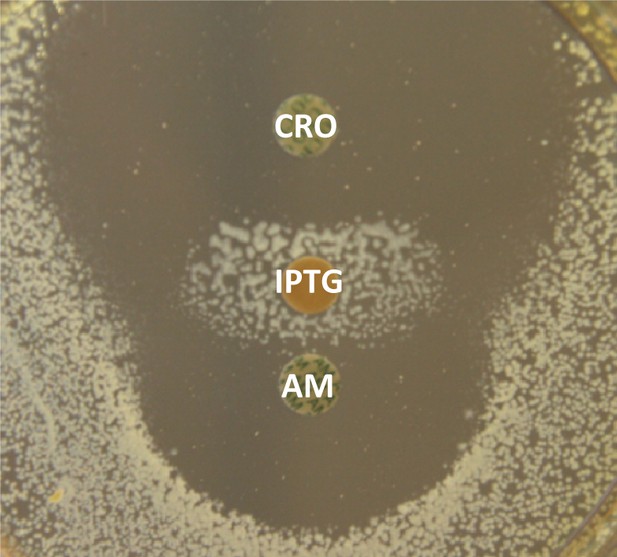
IPTG-inducible expression of β-lactam resistance in mutant M1(pJEH11-1).
The diffusion assay was performed with disks containing 30 µg of ampicillin (AM), 30 µg of ceftriaxone (CRO), or 10 µg of IPTG.
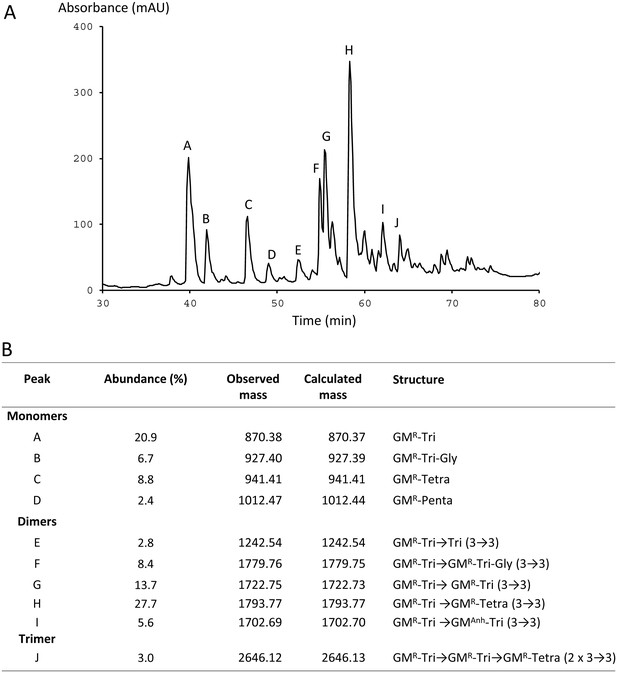
Peptidoglycan composition of mutant M1 grown in presence of ampicillin (16 µg/ml).
(A) rpHPLC profile of disaccharide-peptides. Absorbance was recorded at 210 nm (mAU, absorbance units x 103). (B) Identification of disaccharide-peptides by mass spectrometry. The relative abundance of peptidoglycan fragments was estimated as the percentage of the total integrated area. The observed and calculated monoisotopic mass of muropeptides is indicated in Da. GM, GlcNAc-MurNAc; anh, anhydro; R, reduced; Tri, tripeptide L-Ala-γ-D-Glu-DAP (DAP, diaminopimelic acid); Tetra, tetrapeptide L-Ala-γ-D-Glu-DAP-D-Ala; Tri-Gly, tetrapeptide L-Ala-γ-D-Glu-DAP-Gly; Penta, pentapeptide L-Ala-γ-D-Glu-DAP-D-Ala-D-Ala; 3→3, cross-link generated by L,D-transpeptidation.
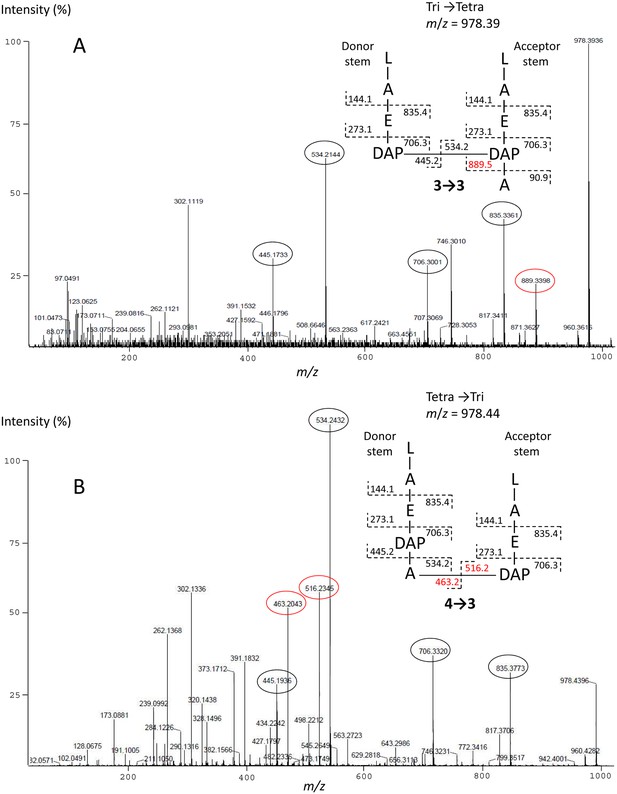
Sequencing of the peptidoglycan cross-link of lactoyl-peptides by tandem mass spectrometry.
The figure illustrates the capacity of the method to discriminate isomers containing 3→3 (A) and 4→3 (B) cross-links. Fragments specific of each isomer are shown in red. L, D-Lac; A, L-Ala or D-Ala; a, C-terminal D-Ala; E, D-Glu; DAP, diaminopimelic acid. All dimers present in peptidoglycan preparations from mutant M1 were identified by this method.
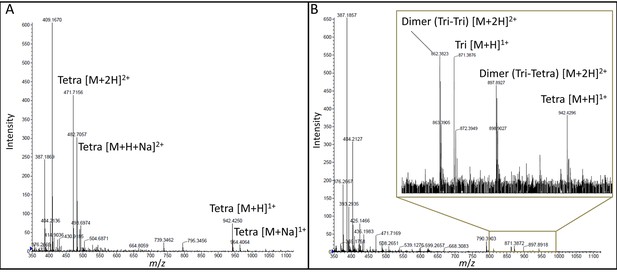
Mass spectrometry analysis of the products of the reactions catalyzed in vitro by YcbB.
(A) Disaccharide-tetrapeptide used as the substrate. The muropeptide GlcNAc-MurNAc-L-Ala1-γ-D-Glu2-DAP3-D-Ala4 (Tetra) was purified from the E. coli cell wall peptidoglycan. The peak at m/z 942.4250 [M+H]1+ corresponds to an observed mass (Mobs) of 941.42 Da in agreement with the calculated mass (Mcal) of 941.41 Da. (B) Reaction products. YcbB (10 µM) was incubated with the disaccharide-tetrapeptide (30 µM) for 2 hr at 37°C revealing the formation of (i) the tripeptide GlcNAc-MurNAc-L-Ala1-γ-D-Glu2-DAP3 (Tri; Mobs = 870.40 Da; Mcal = 870.37 Da) by the L,D-carboxypeptidase activity of YcbB; (ii) the peptidoglycan dimer bis-disaccharide-Tri-Tetra (Mobs = 1793.80 Da; Mcal = 1793.77 Da) by the L,D-transpeptidase activity of YcbB; and the peptidoglycan dimer bis-disaccharide-Tri-Tri (Mobs = 1722.78 Da; Mcal = 1722.73 Da) by the L,D-transpeptidase and L,D-carboxypeptidase activities of YcbB.
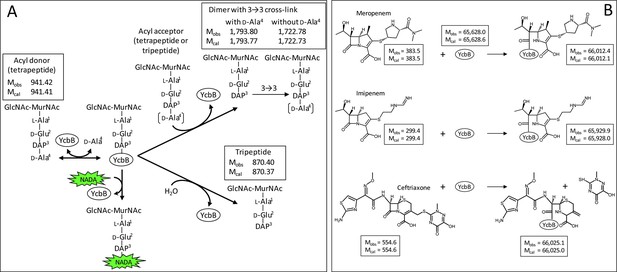
Reactions catalyzed by the L,D-transpeptidase, YcbB.
(A) In vitro cross-linking assay. Incubation of YcbB with the reduced disaccharide GlcNAc-MurNAc-tetrapeptide resulted in the formation of a dimer containing a 3→3 cross-link (L,D-transpeptidase activity). YcbB also hydrolyzed the C-terminal D-Ala4 residue of tetrapeptide stems (L,D-carboxypeptidase activity). The muropeptides were determined by mass spectrometry. The observed (Mobs) and calculated (Mcal) monoisotopic masses are indicated in Daltons. The pentapeptide GlcNAc-MurNAc-L-Ala1-γ-D-Glu2-DAP3-D-Ala4-D-Ala5 was not a substrate of YcbB. The reaction used to label peptidoglycan with a fluorescent derivative of D-Ala (NADA) is indicated. (B) Acylation of YcbB by β-lactams. Incubation of YcbB with two carbapenems, i.e. meropenem and imipenem, led to the acyl-enzymes shown, which were stable. In contrast, the acyl-enzyme formed with ceftriaxone was unstable, accounting for resistance of mutant M1 to this cephalosporin. The observed (Mobs) and calculated (Mcal) average masses are indicated in Daltons. No adduct was observed with amoxicillin.
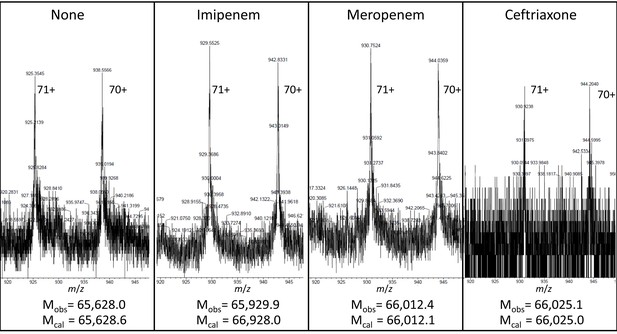
Mass spectrometry analyses of the adducts resulting from acylation of YcbB by the β-lactams imipenem, meropenem, and ceftriaxone.
YcbB (10 µM) was incubated with β-lactams (100 µM) for 1 hr at 37°C. The peaks correspond to the [M+71H]71+ and [M+72H]72+ ions. The observed (Mobs) and calculated (Mcal) masses are indicated. Acylenzymes formed with ceftriaxone (a cephem) were detected although they were slowly hydrolyzed, as previously described for Ldtfm from E. faecium (Triboulet et al., 2013). Acylenzymes formed with ampicillin (a penam) were not detected by mass spectrometry.
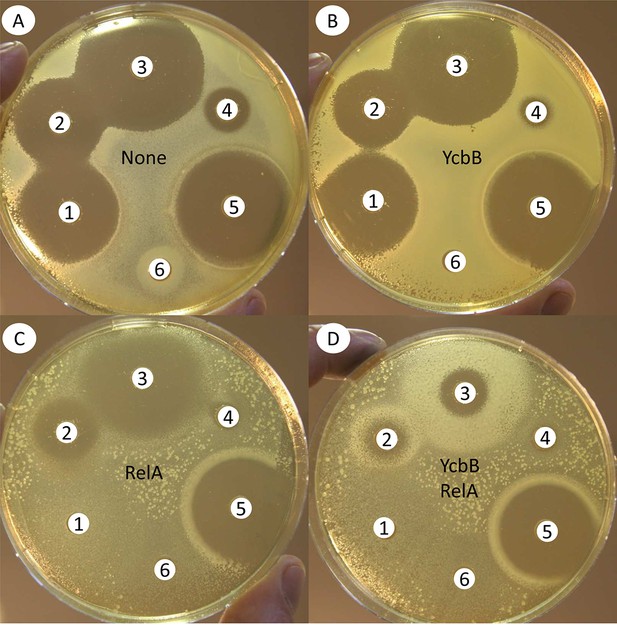
Impact of induction of RelA 1–455 and YcbB synthesis on the activity of β-lactams.
Antibiograms using the disk diffusion assay were performed on BHI agar (A) supplemented with 50 µM IPTG (B), 1% arabinose (C) or both inducers (D), to induce expression of ycbB and relA 1–455 encoded by compatible plasmids pKT2 and pKT8, respectively. Disks were loaded with 10 µg of mecillinam (1), 10 µg of ampicillin (2), 30 µg of ceftriaxone (3), 30 µg of tetracycline (4), 10 µg of imipenem (5), or 30 µg of chloramphenicol (6). Plates were inoculated with BW25113ΔrelA harboring plasmids pKT8(relA) and pKT2(ycbB).
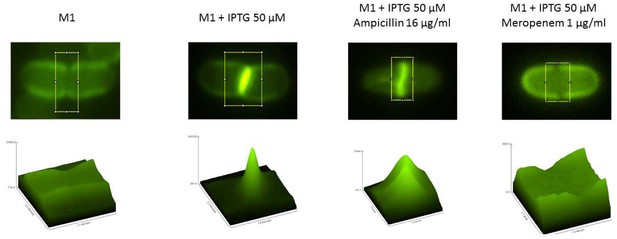
Localization of YcbB activity based on labeling of peptidoglycan with a fluorescent derivative of D-Ala (NADA).
Mutant M1 was grown in the absence or in the presence of 50 µM IPTG to induce ycbB expression. Prior to labeling with NADA, bacteria were incubated with ampicillin, which inhibits PBPs but not YcbB, or meropenem, which inhibits all transpeptidases. The graphics in the lower panel correspond to the surface plot within the yellow rectangle.
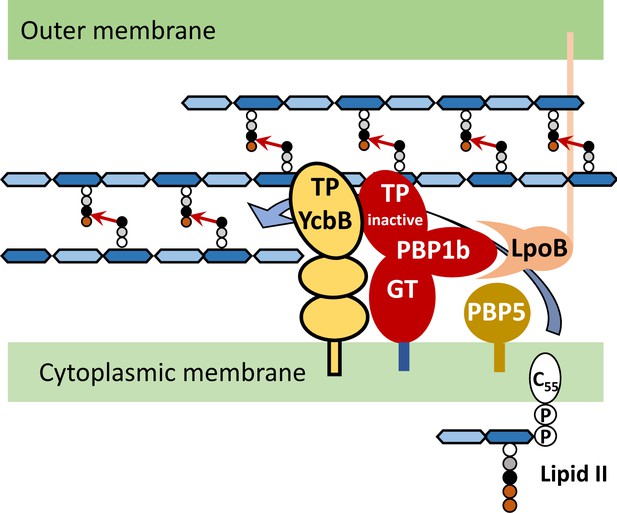
Peptidoglycan polymerization in mutant M1.
The lipid intermediate II (Lipid II) consists in the disaccharide-pentapeptide subunit linked to the undecaprenyl lipid transporter (C55) by a pyrophosphate bond. N-acetylglucosamine linked to N-acetylmuramic acid (MurNAc) by a β1→4 bond is represented by light and dark blue hexagons, respectively. The pentapeptide stem is linked to the D-lactoyl group of MurNAc and assembled by the sequential addition of L-Ala (white circle), D-Glu (grey circle), meso-diamopimelic acid (DAP; black circle), and the dipeptide D-Ala-D-Ala (orange circles). Following translocation through the cytoplasmic membrane, the subunit is polymerized by the glycosyltransferase (GT) activity of PBP1b, which requires binding of LpoB to its UBH2 domain (Vinella et al., 1993). The transpeptidase (TP) activity of PBP1b and other PBPs is bypassed by the TP activity of YcbB that forms 3→3 cross-links (arrows) connecting DAP residues at the 3rd position of stem peptides. The donor substrate of YcbB is generated by the D,D-carboxypeptidase activity of PBP5.
Tables
Susceptibility of E. coli strains determined by the disk diffusion assay.
Antibiotic | Load (µg) | BW25113 | Diameter of inhibition zones (mm) for the indicated strains* | ||||
|---|---|---|---|---|---|---|---|
BW25113Δ4 | M1IPTG 50 µM | M1cured | M1curedpJEH12(ycbB) | M1curedpJEH12(ycbB)IPTG 50 µM | |||
Amoxicillin | 25 | 23 | 24 | 13 | 28 | 28 | 16 |
Ampicillin | 10 | 21 | 21 | ND§ | 27 | 26 | ND |
Amox+Clav† | 20+10 | 20 | 23 | 13 | 27 | 30 | 11 |
Piperacillin | 75 | 29 | 28 | ND | 36 | 36 | ND |
Pip+Tazo‡ | 75+10 | 29 | 30 | ND | 37 | 37 | ND |
Ticarcillin | 75 | 26 | 27 | ND | 37 | 31 | ND |
Mecillinam | 10 | 17 | 22 | ND | ND | ND | ND |
Aztreonam | 30 | 33 | 36 | ND | 49 | 47 | ND |
Cefalotin | 30 | 16 | 18 | ND | 23 | 23 | ND |
Cefoxitin | 30 | 20 | 25 | 29 | 30 | 30 | 30 |
Cefotetan | 30 | 30 | 31 | 25 | 41 | 41 | 27 |
Ceftazidime | 30 | 29 | 30 | ND | 39 | 37 | 9 |
Cefotaxime | 30 | 33 | 36 | ND | 44 | 44 | 9 |
Cefixime | 10 | 28 | 30 | ND | 37 | 38 | ND |
Cefpirome | 30 | 31 | 33 | ND | 41 | 41 | 9 |
Cefoperazone | 30 | 27 | 28 | ND | 41 | 40 | ND |
Moxalactam | 30 | 31 | 32 | 15 | 43 | 40 | 17 |
Ceftriaxone | 30 | 33 | 32 | 15 | 42 | 42 | 18 |
Carbapenems | |||||||
Doripenem | 10 | 31 | 34 | 32 | 38 | 37 | 38 |
Meropenem | 10 | 30 | 34 | 35 | 40 | 41 | 34 |
Imipenem | 10 | 26 | 30 | 35 | 25 | 27 | 28 |
Ertapenem | 10 | 30 | 35 | 37 | 49 | 47 | 37 |
-
*BW25113Δ4 is a derivative of E. coli BW25113 that does not harbor the ynhG, ybiS, erfK, and ycfS genes encoding YcbB paralogues. M1 is a β-lactam-resistant mutant of BW25113Δ4 harboring pJEH11-1(ycbB). M1cured is a derivative of M1 resulting from the spontaneous loss of pJEH11-1(ycbB). Plasmid pJEH12(ycbB) was obtained by replacing the origin of replication (ColE1) and resistance marker (kanamycin) of pJEH11-1(ycbB) by the p15A replication origin and tetracycline resistance marker of plasmid pACY184. The L,D-transpeptidase gene ycbB of pJEH11-1 and pJEH12 are expressed under the control of the IPTG-inducible trc promoter and regulated by the LacI Arg127Leu repressor.
-
†Combination of amoxicillin (20 µg) and clavulanate (10 µg).
-
‡Combination of piperacillin (75 µg) and tazobactam (10 µg).
-
§ND, not detected as the strains grew at the contact of the disk.
Selection of β-lactam-resistant derivatives of E. coli BW25113 harboring various plasmids.
Deletion* | Plasmid 1 | Plasmid 2 | Frequency x 109† |
|---|---|---|---|
None | pACYC184 | pTrc99A | <1 |
None | pJEH12(ycbB) | none | 3200 |
dacA | pJEH12(ycbB) | none | <1 |
dacA | pJEH12(ycbB) | pTrc99AΩdacA | 9100 |
dacB | pJEH12(ycbB) | none | 13,000 |
dacC | pJEH12(ycbB) | none | 9000 |
dacD | pJEH12(ycbB) | none | 9100 |
mrcB | pJEH12(ycbB) | pTrc99A | <1 |
mrcB | pJEH12(ycbB) | pTrc99AΩmrcB | 6400 |
mrcB | pJEH12(ycbB) | pTrc99AΩmrcB TG (E233M) ‡ | <1 |
mrcB | pJEH12(ycbB) | pTrc99AΩmrcB TP (S510A)§ | 1700 |
mrcA | pJEH12(ycbB) | none | 1600 |
pbpC | pJEH12(ycbB) | none | 2700 |
mgtA | pJEH12(ycbB) | none | 2700 |
lpoA | pJEH12(ycbB) | none | 5000 |
lpoB | pJEH12(ycbB) | none | <1 |
-
*Mutants of the Keio collection harboring a kanamycin-resistance gene cassette in place of the indicated gene.
-
†Frequency of survivors obtained by plating 109 colony forming units on agar containing ceftriaxone (32 µg/ml) and IPTG (50 µM). Values are the median from 2 to 8 experiments.
-
‡The glycosyltransferase (GT) module of PBP1b was selectively inactivated by the E233M amino acid substitution.
-
§The transpeptidase (TP) module of PBP1b was selectively inactivated by the S510A substitution.
Selection of β-lactam-resistant derivatives of E. coli CS801-4 harboring various plasmids.
Plasmid 1 | Plasmid 2 | Frequency x 109 | ||||||
|---|---|---|---|---|---|---|---|---|
pACYC184 | pTrc99A | <1 | ||||||
pJEH12(ycbB) | None | <1 | ||||||
pJEH12(ycbB) | pTrc99A | <1 | ||||||
none | pTrc99AΩdacA | <1 | ||||||
pJEH12(ycbB) | pTrc99AΩdacA | 5900 |
-
E. coli CS801-4 harbors deletions of genes pbp4, 5, 6, 7, mrcA, ampH, ampC, and dacD
Mutations detected in M1 to M7.
Mutant | Selection | Position | Mutation | Impact | |||
|---|---|---|---|---|---|---|---|
M1 | Ap | 22,373 | Δ13-nt* | IleRS translation | |||
M2 | Me | 1,960,069 | T→C | I3T in ArgRS | |||
M3 | Me | 1,801,022 | A→C | S517A in ThrRS | |||
M4 | Ap | 2,520,555 | G→T | R40S in GluRS | |||
M5 | Ap | 2,520,540 | C→T | D45N in GluRS | |||
M6 | Ap | 1,948,853 | G→T | T557N in AspRS | |||
M7 | Ap | 1,950,015 | G→A | P170S in AspRS | |||
-
*13-base pair deletion (positions 22,373 to 22,385).
-
Ap, ampicillin; IleRS, isoleucine-tRNA synthetase; Me, mecillinam.
Minimal inhibitory concentration of β-lactams against E. coli strains harboring various plasmids*.
β-lactam | Inducer† | Strains | |||||||
|---|---|---|---|---|---|---|---|---|---|
BW25113 | BW25113pJEH12(ycbB) | M1cured | M1curedpJEH12(ycbB) | BW25113pKT8(relA') | BW25113pKT8(relA')pKT2(ycbB) | BW25113ΔrelApKT8(relA') | BW25113ΔrelApKT8(relA')pKT2(ycbB) | ||
Ampicillin | None | 8 | 8 | 8 | 8 | 8 | 4 | 8 | 4 |
IPTG | 8 | 8 | 8 | 128 | 8 | 4 | 8 | 4 | |
Ara | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 4 | |
IPTG+Ara | 8 | 8 | 8 | 128 | 8 | 64 | 8 | 128 | |
Ceftriaxone | None | 0,05 | 0,05 | 0,05 | 0,05 | 0,05 | 0,05 | 0,05 | 0,05 |
IPTG | 0,05 | 0,05 | 0,05 | 32 | 0,05 | 0,05 | 0,05 | 0,05 | |
Ara | 0,05 | 0,05 | 0,05 | 0,05 | 0,05 | 0,1 | 0,05 | 0,05 | |
IPTG+Ara | 0,05 | 0,05 | 0,05 | 32 | 0,05 | 32 | 0,05 | 32 | |
-
*Minimal inhibitory concentrations were determined by the agar dilution method with an inoculum of 105 colony forming units per spot in brain heart infusion agar after 24 hr of incubation at 37°C. The same results were obtained with 104 and 106 colony forming units, indicating that YcbB in combination with elevated (p)ppGpp rendered the bulk of the population resistant.
-
†Induction was performed with 50 mM IPTG, 1% arabinose (Ara), and a combination of both inducers (IPTG+Ara). IPTG induces expression of the ycbB gene of plasmids pJEH12(ycbB) and pKT2(ycbB). Arabinose induces expression of relA’ encoding the first 455 residues of RelA.
Oligonucleotides used in this study.
Oligonucleotide | Sequence |
|---|---|
PBP1b-inact1 | GAAGAACAGAAAATCGGGCTTTTGCGCCTGAATATTGCGGAGAAAAAGCCATATGAATATCCTCCTTAG |
PBP1b-inact2 | GTTATTTTACCGGATGGCAACTCGCCATCCGGTATTTCACGCTTAGATGGTGTAGGCTGGAGCTGCTTC |
PBP1a-inact1 | GCGCGTTTGTTTATAAACTGCCCAAATGAAACTAAATGGGAAATTTCCACATATGAATATCCTCCTTAG |
PBP1a-inact2 | CAAGTGCACTTTGTCAGCAAACTGAAAAGGCGCCGAAGCGCCTTTTTAAGTGTAGGCTGGAGCTGCTTC |
PBPC-inact1 | ATGCCTCGCTTGTTAACCAAACGCGGCTGCTGGATAACGTTGGCAGCCGCATATGAATATCCTCCTTAG |
PBPC-inact2 | CCGTCAAATGCAGGGTCACGTTGCGCCCGCGTTCAGTTAACGGTTCGCCGTGTAGGCTGGAGCTGCTTC |
MGT-inact1 | GCGGCATTGATAAGCTGGTTTCCCGCGTGCTGGTTCTGGCTGAATGAGTCATATGAATATCCTCCTTAG |
MGT-inact2 | TCGTGAGAGCAAAACGCTGGCCCTCACTTCGCGCGAAGCTTAATCCAGCGTGTAGGCTGGAGCTGCTTC |
PBP1b-NcoI | AAAACCATGGCCGGGAATGACCGCGAGCCAATTGGACGC |
PBP1b-SacI | GTTATGAGCTCGGATGGCAACTCGCCATCCGGTATTTCACGC |
PBP1a-BspHI | TTCTCATGAAGTTCGTAAAGTATTTTTTGATCCTTGC |
PBP1a-SacI | AGCCGGAGCTCGCGTTCACGCCGTATCCGGCATAAACAAGTGCAC |
PMAK-PBP1b | CCAAGGATCCGTAAGGTTGGTTTTCTCCCTCTCCCTGTGGG |
PBP1b-TG1 | GCGACCATGGACCGTCATTTTTACGAGCATGATGGAATC |
PBP1b-TG2 | CGGTCCATGGTCGCCAGCAAAGTATCCACCAGCAAATCC |
PBP1b-TP1 | TCGATTGGCGCCCTTGCAAAACCAGCGACTTATCTGACGGC |
PBP1b-TP2 | TGCAAGGGCGCCAATCGAACGACGCGCCTGCATCGCACGG |
RelA-XbaI | AATCTAGAAATCGATGGTACTTTTCTC |
RelA-PstI | AACTGCAGCTACAGCTGGTAGGTGAACGGC |
DacA-SacI | GGGAGCTCGGCATCTGATGTGTCAAT |
DacA-BamHI | GGGGATCCTTAACCAAACCAGTGATGG |
CDF-for | AACTGCAGTCATGAGCGGATACATAT |
CDF-rev | AAATCGATATCTAGAGCGGTTCAGTAG |





