NADPH oxidase-mediated redox signaling promotes oxidative stress resistance and longevity through memo-1 in C. elegans
Figures
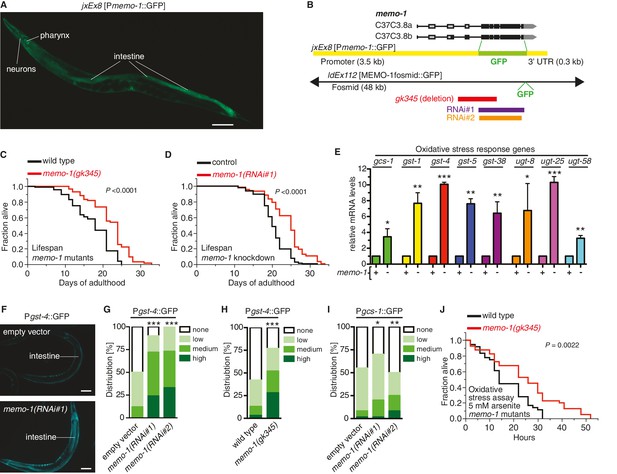
Loss of memo-1 leads to increased ROS.
(A) Transcriptional fusion of the memo-1 promoter (Pmemo-1) with GFP (jxEx8 [Pmemo-1::GFP]) shows that memo-1 is expressed in neurons, pharyngeal cells, and intestine in adult C. elegans. Anterior to the left, ventral side down. Scale bar = 100 µm. (B) Genomic organization of the C37C3.8 (memo-1) locus (gray is untranslated UTR; black are translated exons; adapted from wormbase.org). The memo-1 gene encodes two isoforms (C37C3.8a and C37C3.8b), whereby C37C3.8b is predicted to be 48 amino acids longer than C37C3.8a (297 amino acids). The gk345 allele (red) is a 915 bp deletion. RNAi#1 clone (purple) and RNAi#2 clone (orange) are from Vidal- and Ahringer RNAi libraries, respectively. See Materials and methods for more details. (C) memo-1(gk345) mutants show a 27% increase in mean lifespan compared to wild type (N2) at 20°C. P value determined by log-rank. Statistics and additional lifespan data are in Supplementary file 1. (D) Knockdown of memo-1(RNAi#1) starting on the first day of adulthood in RNAi-sensitive animals (rrf-3(pk1426)) increases mean lifespan by 20% compared to empty RNAi vector control (L4440) at 20°C. P value determined by log-rank. Statistics and additional lifespan data are in Supplementary file 1. (E) memo-1(RNAi#2) treated wild type (N2) (-) have higher mRNA expression levels of the oxidative stress response genes, such as glutamine cysteine synthetase (gcs-1), glutathione-S-transferase (gst-1, 4, 5, 38), and uridine 5'-diphospho-glucuronosyltransferase (ugt-8, 25, 58), compared to empty vector treated wild type (N2) (+), determined by qRT-PCR. 3 replicates of >1000 mixed staged worms per condition were analysed. Data are represented as mean ± s.e.m. P value * <0.05, ** <0.001, *** <0.0001 relative to wild type or control, by one sample t-test, two-tailed, hypothetical mean of 1. (F–I) Loss of memo-1 increases the expression of oxidative stress response genes gst-4 and gcs-1 in the intestine. (F) shows representative pictures of dvIs19 [Pgst-4::GFP] transgenic adult C. elegans treated with empty vector (upper picture; category: none) or memo-1(RNAi#1) (lower picture; category: high). Anterior to the right, ventral side up. Scale bar = 100 µm. (G–I) Quantification of transgenic worms containing the promoter of gst-4 or gcs-1 fused with GFP (dvIs19 [Pgst-4::GFP] and ldIs003 [Pgcs-1::GFP]. Scoring is described in Material and methods. Three trials are shown with N > 60 for each condition and trial. P value by chi2 (* <0.05; ** <0.001; ***p<0.0001). (J) Survival of one-day old adult memo-1(gk345) mutants or wild type (N2) in sodium arsenite (5 mM) was assayed. P value determined by log-rank. Statistics and additional oxidative stress data either with arsenite or tert-butyl hydrogen peroxide are shown in Supplementary file 2.
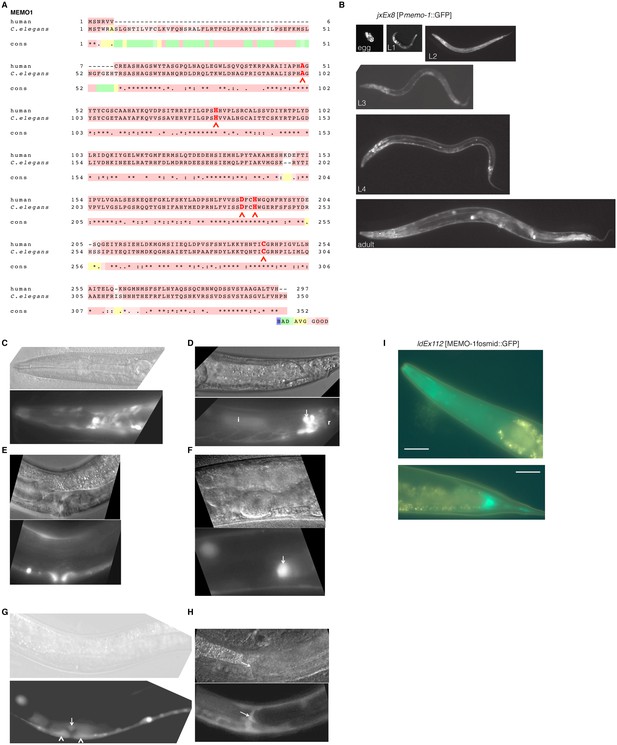
MEMO-1 is a conserved protein that is expressed in many tissues in C. elegans.
(A) Alignment of human Memo1 with C. elegans MEMO-1 amino acid sequence shows high conservation. Human Memo1 Isoform 1 (297 amino acids; Q9Y316 uniprot.org) was aligned with C. elegans memo-1 isoform b (350 amino acids; wormbase.org). 153 out of 297 amino acids (52%) are identical between human and C. elegans. Stars indicate identical amino acids, single dots indicate that size or hydropathy is conserved, and double dots indicate that both size and hydropathy are conserved between the corresponding residues. The amino acids that bind copper are conserved in C. elegans (H49, H81, D189, H192, C244 [MacDonald et al., 2014]) and are indicated with red chevrons. T-coffee was used for the alignment (Notredame et al., 2000). (B–H) memo-1 is expressed in neuronal and non-neuronal cells throughout development and adulthood (B). Promoter memo-1 driven green fluorescent protein (GFP) (jxEx8 [Pmemo-1::GFP]) animals are shown. (C) Pmemo-1::GFP is expressed in some neurons in the head of an L4 wild-type worm, e.g., amphid neurons, including ASJ, and also non-neuronal tissues, such as the pharynx (the procorpus, the anterior bulb, the isthmus and the terminal bulb). (D) In young, adult wild-type worms, Pmemo-1::GFP is expressed in tail neurons, the posterior end of the intestine and the rectal area. Strong Pmemo-1::GFP expression in tail neurons (arrow) and weaker expression in the posterior end of the intestine (i) and the rectal area (r). (E) Pmemo-1::GFP is expressed in the adult vulva. (F) Pmemo-1::GFP is expressed in the spermatheca of wild-type adults. (G) Pmemo-1::GFP is expressed during vulva development at the L4 stage. In an early L4, Pmemo-1::GFP::GFP is expressed weakly around the vulva and expressed strongly in the anchor cell (arrow) and the vulval precursor cells (chevrons). There is also strong expression in the ventral cord neurons. Note that Pmemo-1::GFP is expressed in vulva precursor cells similar to EGFR expression (Haag et al., 2014). (H) Pmemo-1::GFP is expressed in the distal tip cells (arrow). For images (C–H): Top panel shows the Nomarski image and bottom panel the corresponding Pmemo-1::GFP expression. Anterior of the worm is to the left and the ventral side is down; the focus is on the middle plane at 100x magnification. (I) Translational fusion of MEMO-1 with GFP (ldEx112 [MEMO-1fosmid::GFP] is localized to head neurons and pharynx surrounding cells (head; upper picture) and intestinal cells (tail; bottom picture). Scale bar = 20 µm.
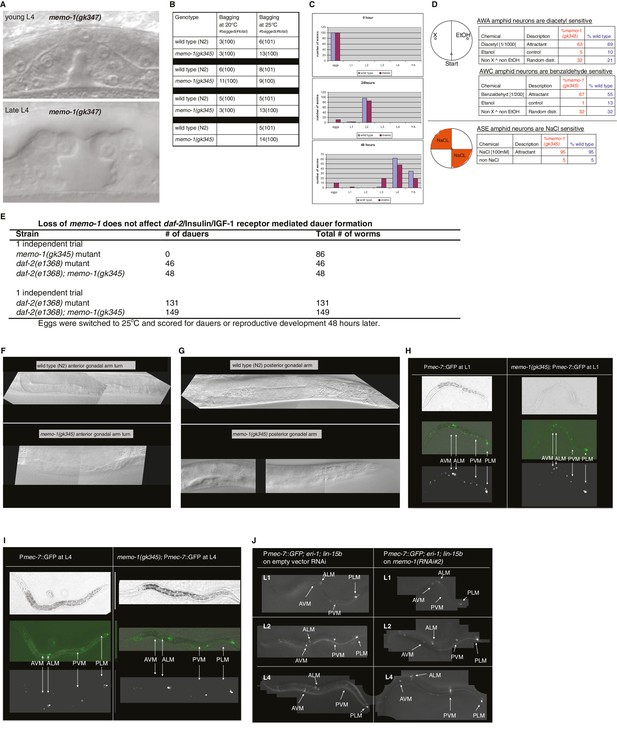
Reverse genetics approach to determine memo-1 function.
(A–B) EGFR phenotypes are not affected by memo-1. EGFR/let-23 mutants fail to form a vulva (Ferguson and Horvitz, 1985) and as a result, EGFR mutants incarcerate their progeny resulting in bags of worms (bagging; [Ferguson and Horvitz, 1985]). The phenotype of the let-23(n1045) mutant has a weakly penetrant multiple vulva (Muv) phenotype at 25°C (Ferguson and Horvitz, 1985). We crossed hermaphrodites of let-23(n1045) that were raised at 20°C either to wild type (N2) or memo-1(gk345) and placed them at 25°C. Three days later, we analyzed the heterozygous progeny for the Muv phenotype at 25°C. The control, which is the heterozygous let-23(n1045)/+; +/+ progeny, had a 27% Muv phenotype at 25°C, whereas the heterozygous let-23(n1045)/+; memo-1(gk345)/+ progeny had a 14% Muv phenotype at 25°C. A t-test showed no significant difference between the two populations for their Muv phenotype (not shown). (A) Developing vulva of memo-1(gk345) mutants looks superficially wild type (wildtype vulva not shown). Top panel shows Normarski image of a young L4 vulva and bottom panel of a late L4 vulva of memo-1(gk345) mutants. (B) memo-1(gk345) mutants do not show increased incidences of bagging. (C) The development speed of memo-1(gk345) mutants is similar to wild type (N > 200; one representative trial out of 3 shown). (D) Top panel schematic: Chemotaxis on agar plates with the triangles indicating the start point for worms. ‘X’ indicates the pole where the compound used in the experiment is placed, and ethanol (EtOH) served as the negative control. 1 µl of sodium azide (NaN3) was added to both poles (green circles) to anesthetize the worms. Lower panel schematic: The agar plates were split into four quadrants. On the surface of the two opposite quadrants 100 mM sodium chloride was added. Tables display results. N > 200 per condition. No statistical difference in chemotaxis was seen between memo-1(gk345) and wild type determined by t-test, unpaired, two-tailed. (E) Loss of memo-1 does not affect insulin/IGF-1 signaling-mediated dauer formation. (F–G) Loss of memo-1 does not affect gonadal migration or morphology at 20°C. (F) Anterior gonadal arm of wild type (upper panel) and memo-1(gk345) mutant (lower panel) at the L4 stage are shown. (G) Posterior gonadal arm of wild type (upper panel) and memo-1(gk345) mutant (lower panel) at the L4 stage are shown. For F-G: anterior of the worm is to the left and the ventral side is down; the focus is on the middle plane at 100x magnification. (H–J) Loss of memo-1 does not affect neuroblast migration. Q neuroblasts and their descendants undergo long-range migration. At the L1 stage the Q neuroblasts divide into QL and QR, which generate some sensory neurons (Sulston and Horvitz, 1977). Both QR and QL descendants start to migrate from the midbody, with the QR descendants migrating anteriorly and the QL descendants migrating posteriorly, where they divide further to give rise to two of the sensory touch receptor neurons (AVM and PVM) (Ch'ng et al., 2003). To test if memo-1 is involved in this migration, we used RNAi to knock down memo-1 or memo-1(gk345) mutants in transgenic muIs32 [Pmec-7::GFP] animals that express GFP in the Q neuroblasts and their descendants (six touch neurons (ALML, ALMR, AVM, PLML, PLMR, and PVM; [Ch'ng et al., 2003]). (H–I) Representative transgenic Pmec-7::GFP animal either in wild-type background (left panel) or memo-1(gk345) mutant background (right panel) before migration of Q neurobasts starts (G) at L1 and after migration (H) at L4. Loss of memo-1 with memo-1(gk345) mutation revealed no deficiency in the migration of the AVM and PVM cells. (J) Representative images of transgenic Pmec-7::GFP animal in an eri-1(mg366); lin- 15b(n744) mutant background treated either with empty vector RNAi (L4440; left panel) or memo-1(RNAi#1 or #2) (right panel; shown is memo-1(RNAi#2)). The eri-1; lin-15b double mutations renders the worm neurons more sensitive to feeding-induced RNAi (Kennedy et al., 2004). Knocking down memo-1 with memo-1(RNAi#1 or #2) did not prevent the migration of the AVM and PVM cells. For all images: anterior of the worm is to the left and the ventral side is down; the focus is on the middle plane at 100x magnification. For (H–I) the same worm is shown per column: top picture is bright field, middle picture is merged GFP with bright field, and bottom picture is GFP channel.
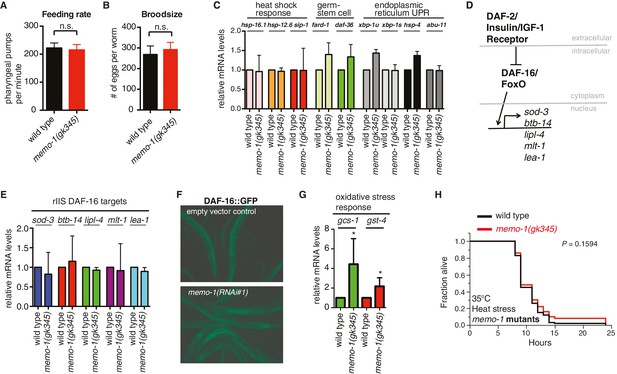
Reduced memo-1 function induces oxidative stress gene expression.
(A) Caloric restriction has been shown to extend lifespan in many species (Kenyon, 2010). Thus, we tested if the memo-1(gk345) mutants are calorically restricted. A genetic model for caloric restriction in the worm has an Eat phenotype, i.e., this worm has a pumping defect in its pharynx, preventing the animal from eating efficiently (Lakowski and Hekimi, 1998). Since Pmemo::GFP is expressed in the pharyngeal area (Figure 1A and Figure 1—figure supplement 1C and l), we analyzed memo-1(gk345) mutants for a pharyngeal pumping defect as a readout of feeding rate. memo-1(gk345) mutants did not have altered pharyngeal pumping compared to wild type (N2) at the L4 stage (N = 10 per strain, P value 0.8747 determined with t-test unpaired two-tailed). (B) Loss of memo-1 does not affect progeny production. Brood size for wild type (N2) (268 ± 15) and for memo-1(gk345) mutants (292 ± 11) was not significantly different. Data represented as mean + s.e.m., P value = 0.1953 determined with t-test, unpaired, two-tailed. (C) Genes involved in the heat shock response, in the germ stem cell-less-mediated longevity (glp-1), and in the unfolded protein response (UPR) are not induced in memo-1(gk345) mutants, as determined by qRT-PCR for the indicated transcripts. N > 200 L4 animals, two merged independent trials in duplicates. xbp-1u = unspliced, xbp-1s = spliced. (D–F) Loss of memo-1 does not affect insulin/IGF-1 signaling. (D) Schematic of insulin/IGF-1 receptor signaling to DAF-16/FOXO in C. elegans. Under normal conditions, insulin/IGF-1 receptor (daf-2) signaling retains the FOXO transcription factor DAF-16 in the cytoplasm. When insulin/IGF-1 signaling is reduced, DAF-16 translocates into the nucleus to initiate transcription of target genes (sod-3, btb-14, lipl-4, mlt-1, lea-1). (E) Downstream targets of DAF-16/Foxo transcription factor that are upregulated when insulin/IGF-1 receptor (daf-2) signaling is reduced (rIIS) are not altered in memo-1(gk345) mutant animals. (F) DAF-16::GFP translocates into the nucleus with daf-2 knockdown (not shown), but not with memo-1 knockdown (N > 60 per condition; zIs356 [DAF-16::GFP]). (G) memo-1(gk345) mutants have higher mRNA levels of oxidative stress response genes gcs-1 and gst-4 compared to wild type (N2), as determined by qRT-PCR. For each condition, two biological samples in duplicates of 200 L4 worms each were analyzed by qRT-PCR. All data are represented as mean ± s.e.m. P values of * <0.05 relative to wild type (N2) control, determined by one sample t-test, two-tailed, hypothetical mean of 1. (H) memo-1 mutants are not heat stress resistant. Day one wild type (N2) and memo-1(gk345) mutants were placed at 35°C and scored every hour for survival (N = 100 per strain). There is no significant difference between wild type (N2) and memo-1(gk345) for heat stress survival determined with log-rank (Mantel-Cox) method to calculate P value = 0.1594.
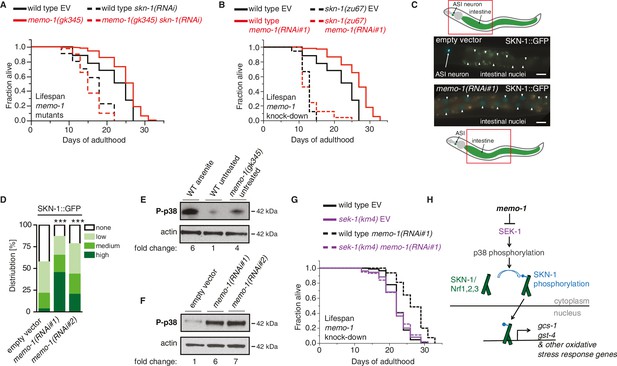
The oxidative stress response pathway is activated and is required for memo-1(-) mediated longevity and oxidative stress resistance.
(A) Knockdown of skn-1 starting on the first day of adulthood abolishes the increased lifespan of memo-1(gk345) mutants. For statistical details and additional lifespans see Supplementary file 1. (B) Knockdown of memo-1(RNAi#1) starting on the first day of adulthood increases lifespan of wild type (N2) animals, but failed to increase lifespan of skn-1(zu67) loss-of-function mutants. For statistical details and additional strains see Supplementary file 1. (C) Schematics and representative pictures of transgenic L4 animals expressing a translational fusion of SKN-1 protein tagged with GFP (ldIs007 [SKN-1::GFP]), treated either with empty vector (upper picture; no SKN-1::GFP in intestine = score none) or memo-1(RNAi#1) (bottom picture; SKN-1::GFP in all intestinal nuclei = score high). Scale bar = 20 µm. Triangles indicate intestinal nuclei. (D) Quantification of SKN-1::GFP in L4 transgenic animals: Knockdown of memo-1(RNAi #1 or #2) starting from the egg stage induced translocation of SKN-1::GFP into the intestinal nuclei. N > 60, three merged trials. *** <0.0001 P values were determined by Chi2 test. Scoring is described in Material and Methods. (E) p38 mitogen-activated protein kinase is phosphorylated (P-p38 MAPK) upon oxidative stress (WT arsenite; 5 mM sodium arsenite for 10 min) or in untreated memo-1(gk345) mutants, compared to untreated wild type (WT untreated). (F) Knockdown of memo-1(RNAi #1 or #2) for two generations led to an increase p38 phosphorylation levels compared to empty RNAi vector control wild type animals. For (E–F) Total extracts from >1000 L4 animals in each condition were analyzed by western for levels of P-p38. Actin was used as a loading control. Fold change indicates the relative P-p38 MAPK to actin levels compared to control (E: WT untreated; F: WT empty RNAi vector control). (G) Knockdown of memo-1(RNAi#1) starting on the first day of adulthood increases lifespan of wild type (N2) animals, but failed to increase lifespan of sek(km4) loss-of-function mutants. For statistical details and additional trials see Supplementary file 1. (H) Schematic of the oxidative stress response pathway in C. elegans. Oxidative stress activates SEK-1/MAPKK phosphorylating p38 MAPK phosphorylating the SKN-1/Nrf transcription factor, promoting its nuclear translocation, which initiates transcription of oxidative stress response genes (e.g., gcs-1 and gst-4).
-
Figure 2—source data 1
Oxidative stress response genes upregulated by loss of memo-1 are transcriptional targets of SKN-1.
- https://doi.org/10.7554/eLife.19493.007
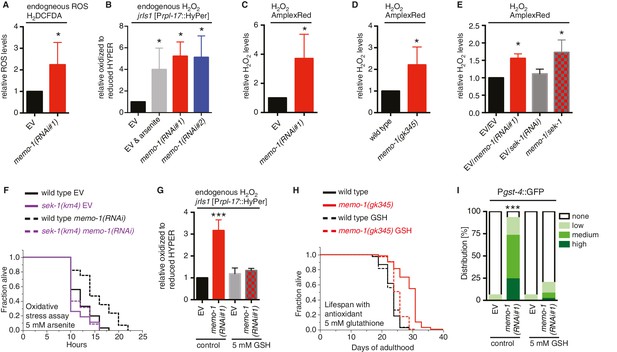
Increased ROS resulting from loss of memo-1 function is required for longevity.
(A) Knockdown of memo-1 increases endogenous ROS levels in vivo, measured using the CM-H2DCFDA fluorescent molecular probe. (B) Knockdown of memo-1 increases endogenous hydrogen peroxide levels, measured in transgenic HyPer worms (jrIs1[Prpl-17::HyPer]), to a similar extent as measured in EV fed animals treated with 5 mM sodium arsenite for 10 min (EV and arsenite). (C–D) Knockdown of memo-1 (C) and memo-1(gk345) mutants (D) have higher hydrogen peroxide levels in vivo compared to wild type (N2) control, as measured with AmplexRed. (E) Knockdown of sek-1 does not suppress the higher hydrogen peroxide levels induced by memo-1(RNAi) measured with AmplexRed. (F) Loss of sek-1 completely suppresses the memo-1(-) mediated oxidative stress resistance. For statistical details and additional trials see Supplementary file 2. (G) Treatment with the antioxidant glutathione (GSH) completely suppresses the higher endogenous hydrogen peroxide levels of transgenic HyPer worms (jrIs1[Prpl-17::HyPer]) treated with memo-1(RNAi) or by measurement with Amplex Red (Figure 3—figure supplement 1C). Eggs were hatched on empty vector (EV) or memo-1(RNAi) food with 5 mM GSH or with solvent (H2O; control) and harvested for assay at larval stage 4 (L4). (H) The antioxidant glutathione (GSH) completely suppresses the longevity of memo-1(gk345) mutants. For statistical details and additional trials with memo-1(RNAi) see Supplementary file 1. (I) The antioxidant glutathione (GSH) completely suppresses the induction of oxidative stress response gene gst-4 in response to memo-1 knockdown. Transgenic animals dvIs19 [Pgst-4::GFP] were placed on empty vector (EV) or memo-1(RNAi) food with 5 mM GSH or solvent (H2O; control) for two generations and day one adults were scored. N > 60, three merged trials. *** <0.0001 P values were determined by Chi2 test. Scoring is described in Material and methods. For (A–C, E) Eggs were hatched on empty vector (EV) or memo-1(RNAi) food and harvested for the assay at larval stage 4 (L4). For (A–E, G) N > 1000 for each condition, three merged trials. All data are represented as mean ± s.e.m. P values * <0.05 relative to wild-type on EV or (D) to wild type (N2), determined by one sample t-test, two-tailed, hypothetical mean of 1.
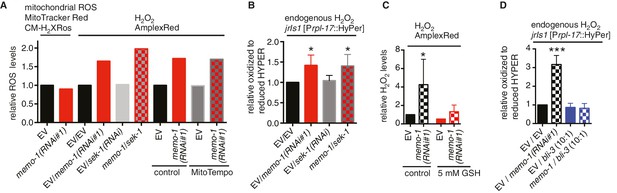
Reduced memo-1 function induces NADPH oxidase specific ROS.
(A) Knockdown of memo-1 does not increase in vivo mitochondrial localized ROS levels as measured with MitoTracker Red CM-H2XRos fluorescent molecular probe. As a control, and run in parallel, memo-1(RNAi) was sufficient to increase hydrogen peroxide measured by Amplex Red, and this was not suppressed by sek-1(RNAi) or by treatment with the mitochondrial specific antioxidant MitoTempo. Eggs were hatched on empty vector (EV) or memo-1(RNAi) (or double RNAi with a ratio 1:1) food and harvested for the assay at larval stage 4 (L4). N > 1000 per condition in this single trial. (B) Knockdown of sek-1 does not suppresse the higher endogenous hydrogen peroxide levels of transgenic HyPer worms (jrIs1[Prpl-17::HyPer]) treated with memo-1(RNAi). (C) The antioxidant glutathione (GSH) completely suppresses the elevated hydrogen peroxide levels of wild-type animals treated with memo-1(RNAi), as measured by Amplex Red. Eggs were hatched on empty vector (EV) or memo-1(RNAi) food in the presence of 5 mM GSH or solvent (control) and harvested for the assay at L4. (D) Knockdown of bli-3/ NADPH oxidase completely suppresses the higher endogenous hydrogen peroxide levels of transgenic HyPer worms (jrIs1[Prpl-17::HyPer]) treated with memo-1(RNAi). A 1:10 ratio for the double RNAi treatment with bli-3 was used. For (J–L) N > 1000 for each condition, three merged trials each with three replicates. All data are represented as mean ± s.e.m. P values * <0.05 and *** <0.0001 relative to wild-type on EV were determined by one sample t-test, two-tailed, hypothetical mean of 1.
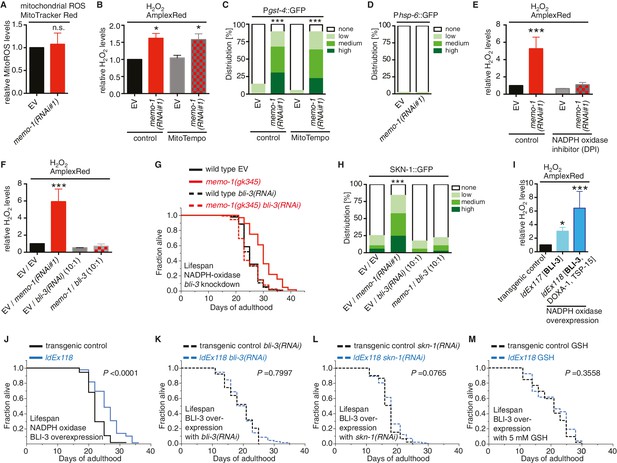
Loss of memo-1 increases ROS via NADPH oxidase to extend lifespan.
(A) Knockdown of memo-1 does not increase mitochondrial localized ROS levels in vivo, measured with MitoTracker Red CM-H2XRos fluorescent molecular probe. In a parallel experiment, the same memo-1(RNAi) treatment was sufficient to increase hydrogen peroxide, as measured by Amplex Red (Figure 3—figure supplement 1A and Figure 4—figure supplement 1). (B) The mitochondrial specific antioxidant MitoTempo did not suppress memo-1(RNAi) induced ROS, measured with Amplex Red. (C) The mitochondrial specific antioxidant MitoTempo did not suppress memo-1(RNAi)) induced expression of gst-4 (dvIs19 [Pgst-4::GFP]). N > 60, three merged trials. *** <0.0001 P values were determined by Chi2 test. Scoring is described in Material and methods. (D) The mitochondrial stress response gene hsp-6 (zcIs13 [Phsp-6::GFP]) was not induced by memo-1(RNAi). N > 100 for each condition, three merged trials. As a control and run in parallel, memo-1(RNAi) was sufficient to increase Pgst-4::GFP expression (Figure 5—figure supplement 1B). (E) A short (15 min) treatment with the NADPH oxidase inhibitor Diphenyleneiodonium (DPI) completely suppressed the elevated hydrogen peroxide levels of wild-type worms treated with memo-1(RNAi). (F) Knockdown of bli-3/ NADPH oxidase completely suppressed the elevated hydrogen peroxide levels of wild-type worms treated with memo-1(RNAi). As reported in Chávez et al. (2009), a 1:10 ratio for the double RNAi treatment with bli-3 was used because of the strong bli-3 blistering phenotype. For corresponding experiments with transgenic HyPer worms (jrIs1[Prpl-17::HyPer]), see Figure 3—figure supplement 1D. (G) Adulthood specific knockdown of bli-3/ NADPH oxidase (undiluted RNAi) completely suppressed the longevity of memo-1(gk345) mutants. For statistical details and additional trials see Supplementary file 1. (H) Knockdown of bli-3/ NADPH oxidase completely suppressed memo-1(RNAi) induced nuclear translocation of SKN-1 protein (ldIs007 [SKN-1::GFP]). N > 60, three merged trials. *** <0.0001 P values were determined by Chi2 test. Scoring is described in Material and methods. (I) Transgenic worms overexpressing BLI-3 (ldEx117) alone, or triple transgenic worms (ldEx118) overexpressing BLI-3 and co-factors for NADPH oxidase complex maturation and stability (DOXA-1 and TSP-15; [Moribe et al., 2012]) showed an increased hydrogen peroxide level compared to control transgenic animals. Mixed stage worms, N > 1000 for each condition, three merged trials. All data are represented as mean ± s.e.m. P values * <0.05 or *** <0.0001 relative to transgenic control (ldEx102) were determined by one sample t-test, two-tailed, hypothetical mean of 1. (J–M) Triple transgenic worms (ldEx118) overexpressing BLI-3, DOXA-1, TSP-15 showed an increased lifespan compared to transgenic control (ldEx102). The increased lifespan was dependent on bli-3 (K), skn-1 (L), and ROS (M). (J–M) are from the same trial (Supplementary file 1). P-value determined by log-rank. Overexpression of BLI-3 alone (ldEx117) was also sufficient to increase lifespan (Supplementary file 1). For statistical details and additional trials see Supplementary file 1. For (A, B, E, F) eggs were hatched on empty vector (EV) or memo-1(RNAi) (or double RNAi with a ratio 1:1, except where indicated) food, and harvested for assay at larval stage 4 (L4). N > 1000 for each condition, three merged trials. All data are represented as mean ± s.e.m. P values * <0.05 or *** <0.0001 relative to wild-type on EV were determined by one sample t-test, two-tailed, hypothetical mean of 1.
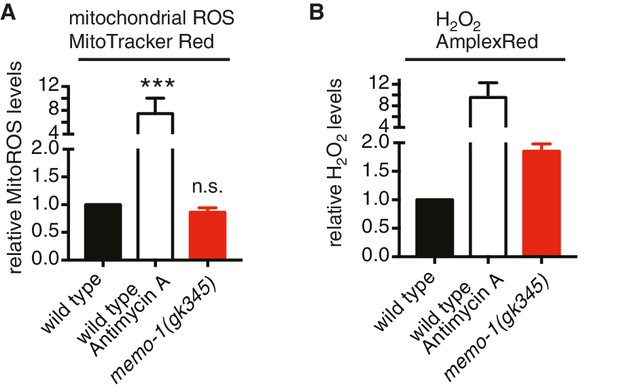
Loss of memo-1 does not increase mitochondrial ROS.
(A) Loss of memo-1 does not increase in vivo mitochondrial localized ROS levels as measured with MitoTracker Red CM-H2XRos fluorescent molecular probe (five independent trials). To induce mitochondrial ROS, Antimycin A treatment on wild type was used as a control. (B) For two out of the five biological independent trials shown in (A), animal populations were split in half and the corresponding half was used to measure hydrogen peroxide in the supernatant with Amplex Red, in parallel to the mitochondrial ROS measurements with MitoTracker Red CMXRos. Mutants that lack memo-1 showed an almost 2-fold increase in ROS measured with Amplex Red, but showed no significant difference in mitochondrial ROS compared to wild type measured with MitoTracker Red CM-H2XRos fluorescent molecular probe (A). For (A–B) N > 1000 per condition per trial. All data are represented as mean ± s.e.m. P values *** <0.0001 relative to wild type were determined by one sample t-test, two-tailed, hypothetical mean of 1. Note that (B) are only two independent trails and therefore fail any statistical significance.
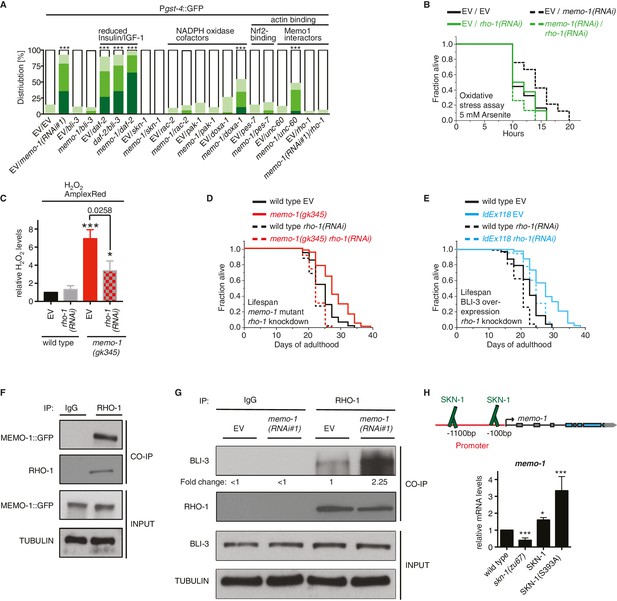
MEMO-1 inhibits NADPH oxidase via RHO-1/GTPase.
(A) Summary of the targeted RNAi screen using the SKN-1 target gene gst-4 expression (dvIs19 [Pgst-4::GFP]) as a read-out for memo-1(-) induced NADPH oxidase activity. N > 60, three merged trials. *** <0.0001 P values were determined by Chi2 test. For individual trials see Figure 5—figure supplement 1. Scoring is described in Material and methods. (B) Loss of rho-1 completely suppresses the memo-1(-) mediated oxidative stress resistance. For statistical details see Supplementary file 2. (C) Knockdown of rho-1 lowered the memo-1(-) mediated ROS induction in all three independent biological trials. Eggs were hatched on empty vector (EV) or rho-1(RNAi). All data are represented as mean ± s.e.m. P values * <0.05 and *** <0.0001 relative to wild type EV control were determined by one sample t-test, two-tailed, hypothetical mean of 1. The memo-1(gk345) mutants treated with rho-1(RNAi) was compared to the memo-1(gk345) mutants treated with EV using paired t-test, one-tailed. (D) The longevity of memo-1(gk345) mutants was completely suppressed by adulthood specific knockdown of rho-1. For statistical details see Supplementary file 1. (E) The longevity of triple transgenic worms (ldEx118) overexpressing BLI-3, DOXA-1, TSP-15 was partially suppressed by adulthood specific knockdown of rho-1. For statistical details see Supplementary file 1. (F) MEMO-1 physically interacts with RHO-1 under wild-type conditions. Lysates from MEMO-1::GFP expressing animals ldEx112 [MEMOfosmid::GFP] were subjected to immunoprecipitations with a C. elegans specific RHO-1 antiserum and immunoblotted with a GFP antibody following SDS-PAGE. (G) Knockdown of memo-1 enhances the physical interaction of RHO-1 with BLI-3. Endogenous RHO-1 was immunoprecipited from lysates of wild type worms fed either EV or memo-1(RNAi) using RHO-1 antibody and was immunoblotted with BLI-3 antiserum. The relative BLI-3 levels in Co-IP samples to input samples is expressed as a fold change of memo-1(RNAi) to the empty vector control (EV). (H) Feedback loop: The memo-1 promoter contains four predicted SKN-1 binding sites (wormbase.org). In a genome-wide screen, transgenic SKN-1::GFP showed a high signal on two skn-1-binding sites in the memo-1 promoter region (upper panel; [Niu et al., 2011]). Lower panel shows that the memo-1 mRNA levels are lower in skn-1(zu67) loss-of-function mutants and higher in conditions of SKN-1 overexpression (ldIs007 [SKN-1::GFP]) and constitutively nuclear SKN-1 overexpression (ldIs020 [SKN-1S393::GFP]; [Tullet et al., 2008]) in transgenic animals, compared to wild type (N2), determined by qRT-PCR. Three biological samples of each 100 L4 worms per strain per trial. Data are represented as mean ± s.e.m. P value * <0.05, ** <0.001, *** <0.0001 relative to wild type or control, by one sample t-test, two-tailed, hypothetical mean of 1.
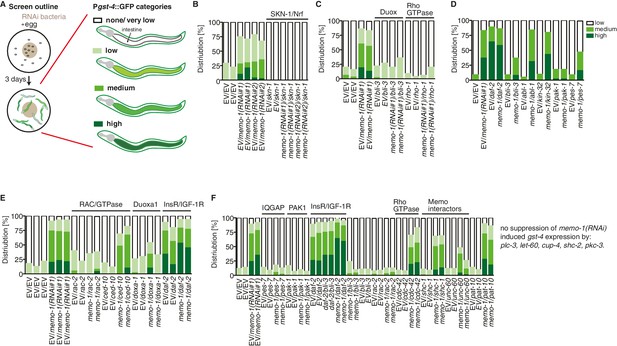
Targeted RNAi screen to discover genes mediating memo-1 activation of SKN-1.
(A) Schematic outline of the targeted RNAi screen. Eggs were placed on RNAi food and early adults (left panel) were scored for their gst-4 (dvIs19 [Pgst-4::GFP]) induction in the intestine (right panel). Scoring is described in Material and methods. We screened more than 200 genes; the positive hits were retested and results are represented in the graphs. For (B–F): each graph shows 1 trial with 2–3 replicates with N > 60 for each condition and replicate. RNAi clones were sequenced to confirm their identity. Mammalian homologues or the gene family are indicated. (B) skn-1(RNAi) completely abolishes gst-4 expression. (C) The dual oxidase (Duox) BLI-3 and RHO-1 (Rho GTPase), but not CDC-42 (Rho GTPase; see F) are required for memo-1(RNAi) induced gst-4 expression. (D) The p21-activated kinase PAK-1 is required for memo-1(RNAi) induced gst-4 expression. (E) The Duoxa1 (Duox maturation factor) doxa-1, rac-2 (RAC/GTPase), but not ced-10 (RAC/GTPase) are required for memo-1(RNAi) induced gst-4 expression. Reducing daf-2/insulin/IGF-1 receptor (InsR/IGF-1R) signaling induces gst-4 expression and this is additive to memo-1(RNAi) induced gst-4 expression. (F) pes-7/IQGAP is completely and unc-60/cofilin is partially required for memo-1(RNAi) induced gst-4 expression. Importantly, bli-3(RNAi) does not block gst-4 expression per se, since daf-2(RNAi)-mediated gst-4 expression was still occurring even when bli-3 was knocked down. Several potential interesting candidates were not required for memo-1(RNAi) induced gst-4 expression, including shc-1, shc-2, plc-3, let-60, cup-4, pkc-3.
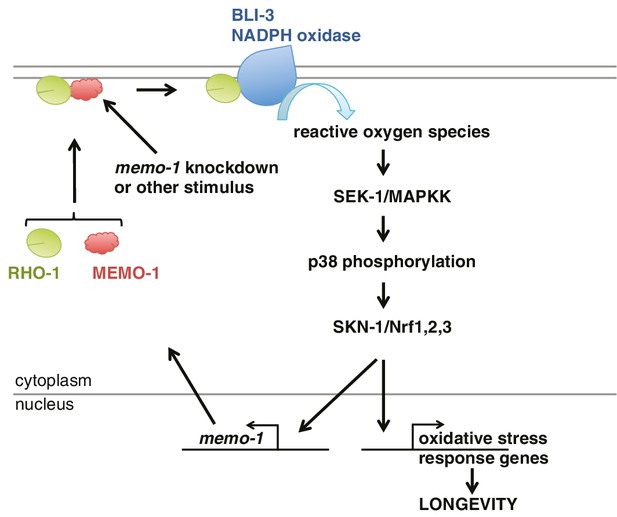
Model of how MEMO-1 activates NADPH oxidase activity to promote oxidative stress resistance and longevity.
Under normal conditions, MEMO-1 is complexed with RHO-1/GTPase. Loss of memo-1 frees RHO-1 to enhance BLI-3/NADPH oxidase activity to generate localized ROS, which activates p38 MAPK signaling to SKN-1 to transcribe genes important for oxidative stress resistance and longevity. Similar to ERBB2 recruiting Memo1 in breast cancer cells (Marone et al., 2004), we speculate that in C. elegans a stimulus might activate a cell-surface receptor to recruit MEMO-1, thereby freeing RHO-1 to promote BLI-3/NADPH oxidase activity. Because SKN-1 also transcribes memo-1 (Figure 5H) resulting in a negative feedback loop to shut off BLI-3/NADPH oxidase activity, memo-1 RNAi or mutation breaks this feedback loop, resulting in continuously enhanced BLI-3 activity.
Additional files
-
Supplementary file 1
Loss of memo-1 increases adult lifespan.
- https://doi.org/10.7554/eLife.19493.015
-
Supplementary file 2
Loss of memo-1 increases oxidative stress resistance.
- https://doi.org/10.7554/eLife.19493.016
-
Supplementary file 3
Primer Sequences.
- https://doi.org/10.7554/eLife.19493.017





