Dormancy-specific imprinting underlies maternal inheritance of seed dormancy in Arabidopsis thaliana
Figures
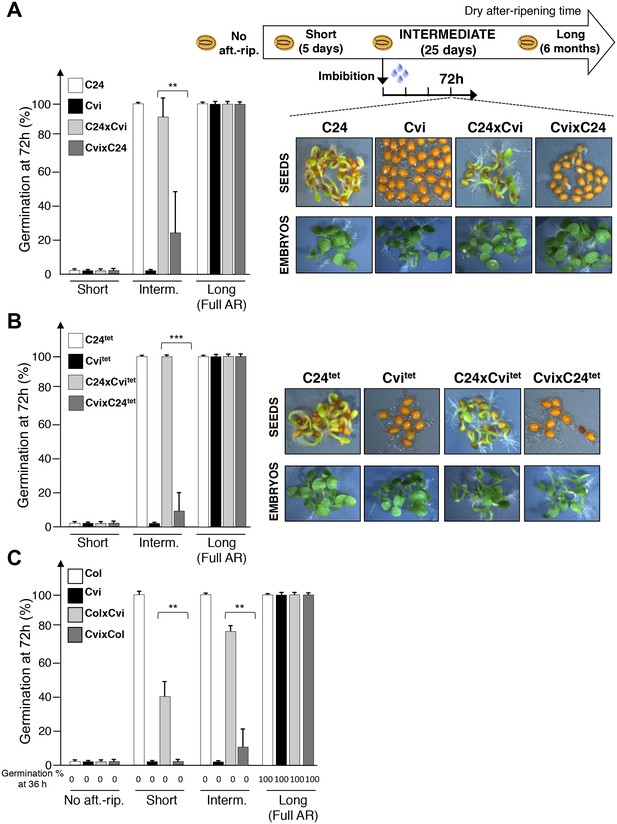
Maternal inheritance of seed dormancy.
(A) Histograms depicting percent germination of C24, Cvi, C24xCvi F1 and CvixC24 F1 seed populations after-ripened for short (five days), intermediate (25 days) or long (six months) time periods. Germination was scored after 72 hr of seed imbibition (4 replicates, n = 150–200, **p<0.01, ***p<0.001, Student’s t test). Pictures show intermediately after-ripened C24, Cvi, C24xCvi F1 and CvixC24 F1 seeds 72 hr after imbibition (upper panel) and the respective embryos (lower panel) following testa and endosperm removal 4 hr after seed imbibition. (B) Same as (A) using C24tet, Cvitet, C24xCvitet F1 and CvixC24tet F1 seeds. (C) Same as (A) with Col, Cvi, ColxCvi F1 and CvixCol F1 seeds that were not after-ripened (No aft.-rip) or after-ripened for short (10 days), intermediate (20 days) and long (six months) time periods.
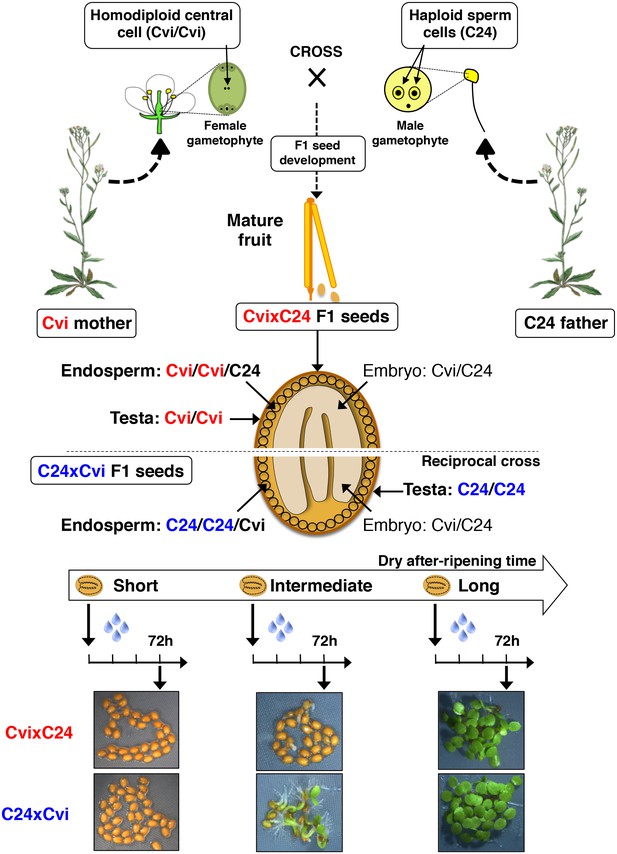
Procedure for assessing CvixC24 F1 and C24xCvi F1 hybrid seed production, dry after-ripening and seed germination.
A Cvi mother plant pollinated with C24 pollen produces CvixC24 F1 seeds, whose endosperm cells bear two Cvi maternal genomes and one C24 paternal genome. C24xCvi F1 seeds are obtained by performing the reciprocal cross. CvixC24 and C24xCvi embryo cells bear one Cvi genome and one C24 genome. The testa is composed of dead integumentary maternal tissue, which is homodiploid Cvi (Cvi/Cvi) and C24 (C24/C24) in CvixC24 and C24xCvi seeds, respectively. Freshly harvested F1 hybrid seeds are dry after-ripened for short (five days), intermediate (25 days) or long (six months) time periods. At the end of each after-ripening period, F1 hybrids seeds are imbibed and germination is scored 72 hr after imbibition.
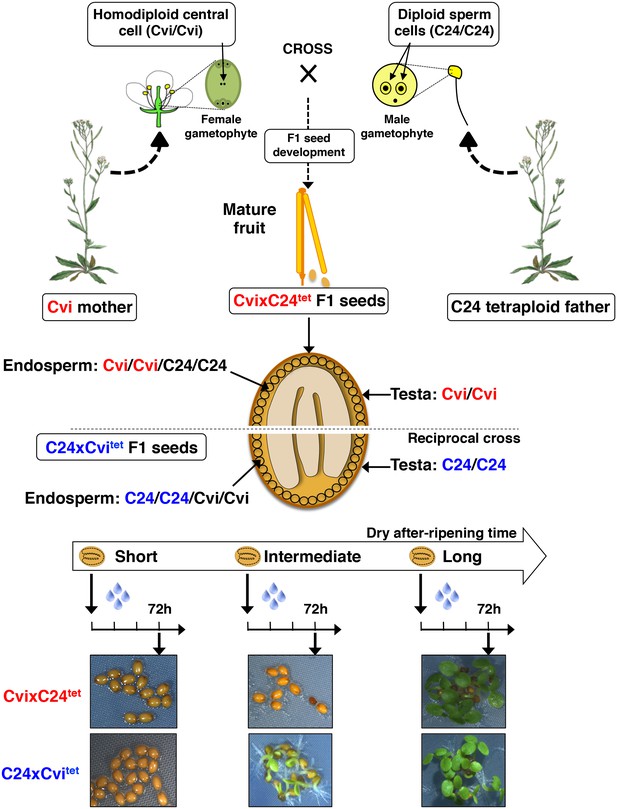
Procedure for the assessment of CvixC24tet F1 and C24xCvitet F1 hybrid seed production, dry after-ripening and seed germination.
Same as Figure 1—figure supplement 1 except that CvixC24tet and C24tet are used as pollen donors. The resulting CvixC24tet F1 hybrid seeds have endosperm cells bearing two Cvi maternal genomes and two C24 paternal genomes, whereas and C24xCvitet F1 hybrid seeds have endosperm cells bearing two C24 maternal genomes and two Cvi paternal genomes.
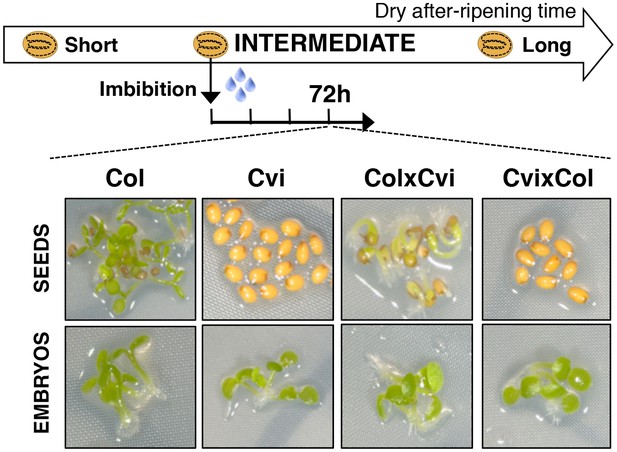
Maternal inheritance of seed dormancy.
Pictures show intermediately after-ripened Col, Cvi, ColxCvi F1 and CvixCol F1 seeds 72 hr after imbibition (upper panel) and the respective embryos (lower panel) following endosperm removal 4 hr after seed imbibition.
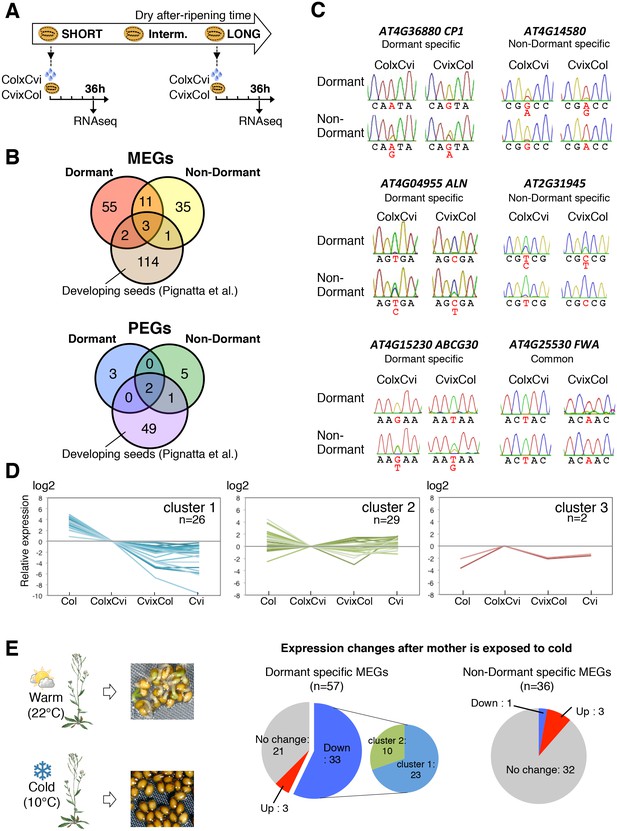
Dynamic changes in imprinted gene expression.
(A) Seed treatment procedure used to isolate endosperm from seeds. CvixCol and ColxCvi F1 seeds were after-ripened for short (10 days) or long (two months) time periods. RNA that was extracted from endosperm dissected 36 hr after imbibition was processed for RNA-seq and RT-PCR for Sanger sequencing analysis. (B) Overlap among maternally expressed imprinted genes (MEGs) and paternally expressed imprinted genes (PEGs) in the endosperm of dormant and non-dormant seeds and of developing seeds according to Pignatta et al. (2014). (C) Sanger sequencing chromatograms at SNPs of selected genes. RNA from shortly after-ripened (Dormant) and fully after-ripened (Non-Dormant) CvixCol and ColxCvi endosperms were analyzed. Three Dormant-specific MEGs and two Non-dormant-specific MEGs and one MEG found in all data sets (Dormant, Non-dormant, and Early embryogenesis) are shown. Nucleotides at SNP sites are highlighted in red. (D) K-means clusters of dormant-specific MEGs expression levels. RNA samples extracted from endosperms of shortly after-ripened Col, ColxCvi, CvixCol, and Cvi seeds were analyzed. For each gene, expression levels relative to those found in ColxCvi endosperms are shown. (E) Gene expression profiles in response to cold treatment of the mother plants during seed development. Left panel: Mother plants exposed to cold temperature produce highly dormant seeds. Right panel: The pie charts group MEGs according to how their expression responds upon seed imbibition after the mother plant was exposed to cold temperatures. 'Down' means that expression was downregulated more than two fold (p < 0.05), 'Up' means that expression was upregulated more than two fold (p < 0.05).
-
Figure 2—source data 1
Genes that are differentially expressed between the endosperm+testa and endosperm samples.
- https://doi.org/10.7554/eLife.19573.008
-
Figure 2—source data 2
mRNA expression of MEGs in the testa+endosperm and endosperm samples.
- https://doi.org/10.7554/eLife.19573.009
-
Figure 2—source data 3
Imprinted genes in the endosperm of mature seed.
- https://doi.org/10.7554/eLife.19573.010
-
Figure 2—source data 4
mRNA expression of dormancy-specific MEGs.
- https://doi.org/10.7554/eLife.19573.011
-
Figure 2—source data 5
Expression of MEGs in response to cold treatment during seed development.
- https://doi.org/10.7554/eLife.19573.012
-
Figure 2—source data 6
RNA-seq data for all genes for imprinting in the endosperm of mature seed.
- https://doi.org/10.7554/eLife.19573.013
-
Figure 2—source data 7
Resequencing of amplicons containing SNPs.
- https://doi.org/10.7554/eLife.19573.014
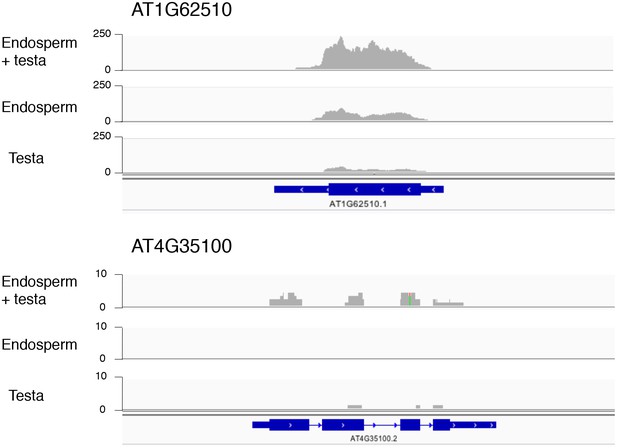
Histograms depicting the read coverage for the AT1G62510 and AT4G35100 genes.
These genes have higher expression in the endosperm + testa RNA sample relative to the endosperm RNA sample. The distribution of RNA-seq reads across the gene (schematized in blue) for each sample (endosperm + testa, endosperm and testa) are shown. Numbers indicate the read depth range (AT1G62510 — 0–250; AT4G35100 — 0–10).
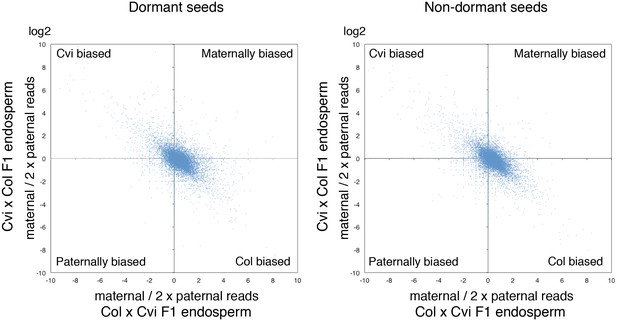
Ratio of maternal reads to duplicated paternal reads for endospermic mRNA isolated from CvixCol and ColxCvi F1.
The charts display the log values of the ratio of maternal to duplicated paternal reads for each gene (blue dots).

Sanger sequencing chromatograms at SNPs of dormant-specific MEGs.
RNA extracted from shortly after-ripened (Dormant) and fully after-ripened (Non-Dormant) CvixCol and ColxCvi endosperm was analyzed. Nucleotides at SNP sites are highlighted with red.
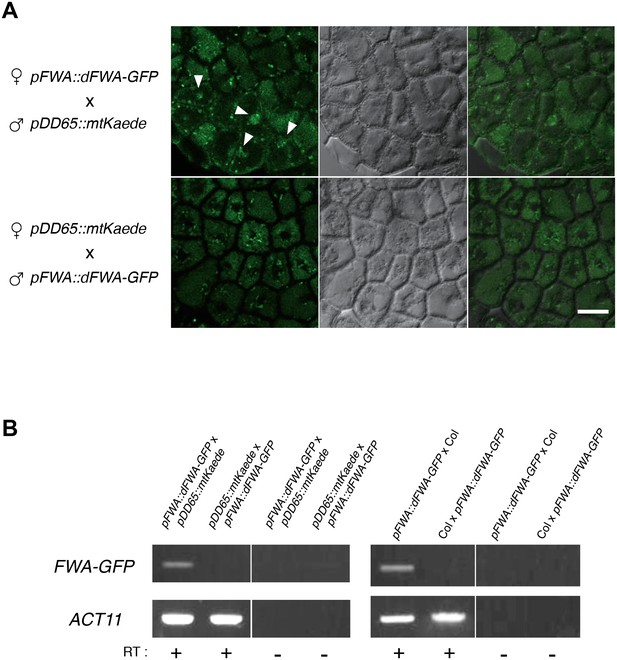
Imprinting of FWA is observed in the endosperm of mature seed.
(A) Confocal images showing FWA-GFP localization in the mature endosperm of the reciprocally crossed F1 seeds. The images were made using confocal microscopy of female pFWA::dFWA-GFP x male pDD65::mtKaede (upper panels) and female pDD65::mtKaede x male pFWA::dFWA-GFP (lower panels) endosperms. Endosperm tissue autofluorescence and FWA-GFP fluorescence signals were captured through the band-pass filter for GFP (left panels). The endosperm cells were visualized by DIC optic (middle panels), and both images were merged (right panels). Although broad background fluorescence were visible in the cytosol in both directions of cross, some cells showed nuclear-localized green fluorescence that is only detectable in the female pFWA::dFWA-GFP cross. pDD65::mtKaede signals distributed with a speckle-like pattern (that probably corresponds to the distribution of mitochondria) in the cytosolic space of all cells. The low signal intensity might be due to the low expression level of FWA in mature seeds, especially in the Col-0 background (Figure 2—source data 3). Bar = 20 μm. (B) RT-PCR detecting the FWA-GFP transgenic mRNA in the mature endosperm of F1 seeds. RNA extracted from endosperm dissected 36 hr after imbibition was analyzed by RT-PCR. FWA-GFP cDNA was detected in the endosperm of pFWA::dFWA-GFP x pDD65::mtKaede and pFWA::dFWA-GFP x Col F1 seeds but not in that of pDD65::mtKaede x pFWA::dFWA-GFP and Col x pFWA::dFWA-GFP F1 seeds. ACT11 is used as a positive control.
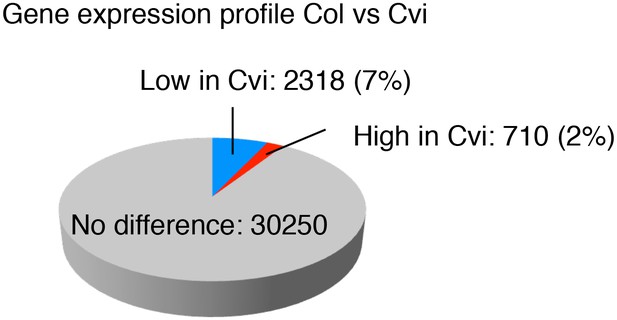
Comparison of the gene expression profiles of Col and Cvi.
Pie chart showing the proportion of differentially regulated genes (more than two-fold, p<0.05) in endosperms of mature seeds of Col and Cvi upon imbibition.

Testa removal does not trigger the germination of highly dormant seeds that developed under low temperatures (10°C).
https://doi.org/10.7554/eLife.19573.020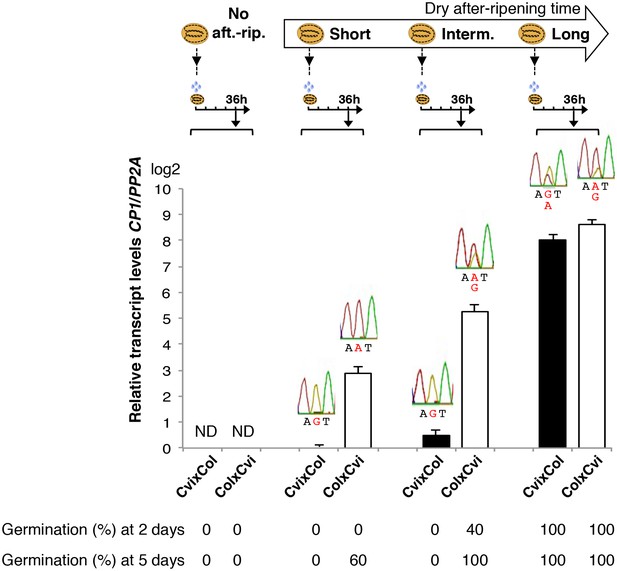
Low and preferential maternal CP1 allele expression is prolonged in dormant seeds but not in non-dormant seeds.
CvixCol F1 and ColxCvi F1 seeds were either not after-ripened (No aft.-rip.) or after-ripened for short (10 days), intermediate (20 days), and long (two months) time periods. Percent germination 2 and 5 days after seed imbibition is shown. RNA extracted from endosperm 36 hr after seed imbibition, i.e. prior to germination, was used for qPCR and RT-PCR for Sanger sequencing analysis. CP1 expression levels relative to those found in the endosperm from CvixCol F1 seeds that had been after-ripened for an indicated time period are shown. CP1 expression levels were normalized to those of PP2A. ND: no detection.

Low and preferential maternal CP1 alelle expression is prolonged in dormant seeds but not in non-dormant seeds.
CvixC24 F1 and C24xCvi F1 seeds were after-ripened for an intermediate time period (25 days). RNA extracted from seed coats 48 hr and 72 hr after seed imbibition, i.e. prior to germination, was used for qPCR and RT-PCR for Sanger sequencing analysis. Expression levels of CP1 were normalized to those of PP2A. CP1 expression levels relative to those found in CvixC24 F1 endosperms 48 hr and 72 hr after imbibition are shown. Pictures show C24xCvi F1 and CvixC24 F1 seeds that had been after-ripened for an intermediate time period (25 days) five days after imbibition.
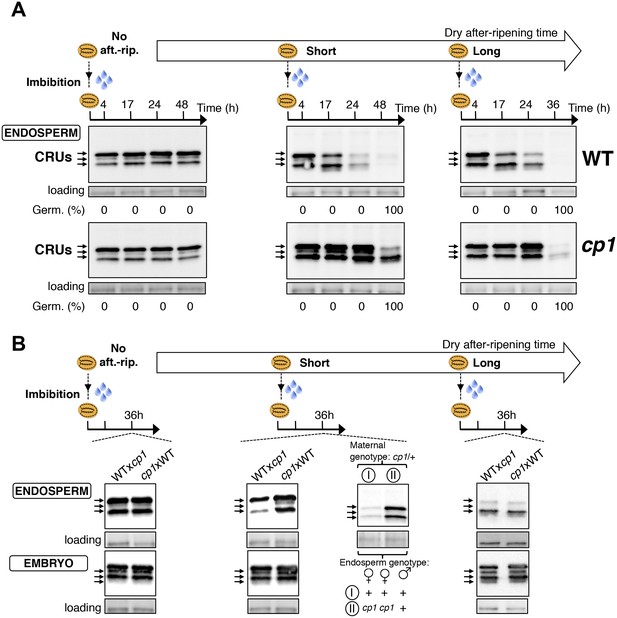
CP1 promotes the decay of CRU proteins.
CRU protein decay is controlled through CP1 maternal gametophytic alleles. (A) Endosperms from non-after-ripened (No aft.-rip.) WT and cp1 seeds or from WT and cp1 seeds after-ripened for short (three days) or long (two months) time periods were dissected at the indicated times after seed imbibition. Proteins isolated from the same number of WT (Col) or cp1 endosperms were separated on SDS PAGE gels and probed with 12S antibody serum. The specificity of the antibody was confirmed using protein extracts isolated from the endosperm and embryo of the cruabc mutant (Figure 4—figure supplement 1). For a given after-ripening time, WT and cp1 endosperm proteins were separated on the same SDS PAGE gel. An aspecific band is used as a loading control. Percent germination at each time-point after seed imbibition is indicated. Arrows indicate highly abundant cruciferin (CRU) proteins. (B) WTxcp1 F1 and cp1xWT F1 seeds (obtained by reciprocally crossing WT (Col) and cp1 plants) were subject to dry after-ripening treatments as in (A). For each after-ripening treatment, endosperms and embryos were dissected 36 hr after seed imbibition. Proteins isolated from the same number of endosperms or embryos were processed as in (A). Seeds obtained after crossing cp1/+ heterozygous mother plants with Col WT (+/+) pollen were after-ripened for a short time period (three days). Endosperms were dissected 36 hr after imbibition, whereas embryos were further cultured for later genotyping in order to distinguish endosperms according to their (i) +/+/+ or (ii) cp1/cp1/+ genotype. Endosperms with the same genotype were pooled. Proteins extracted from the two endosperm pools were processed as in (A).
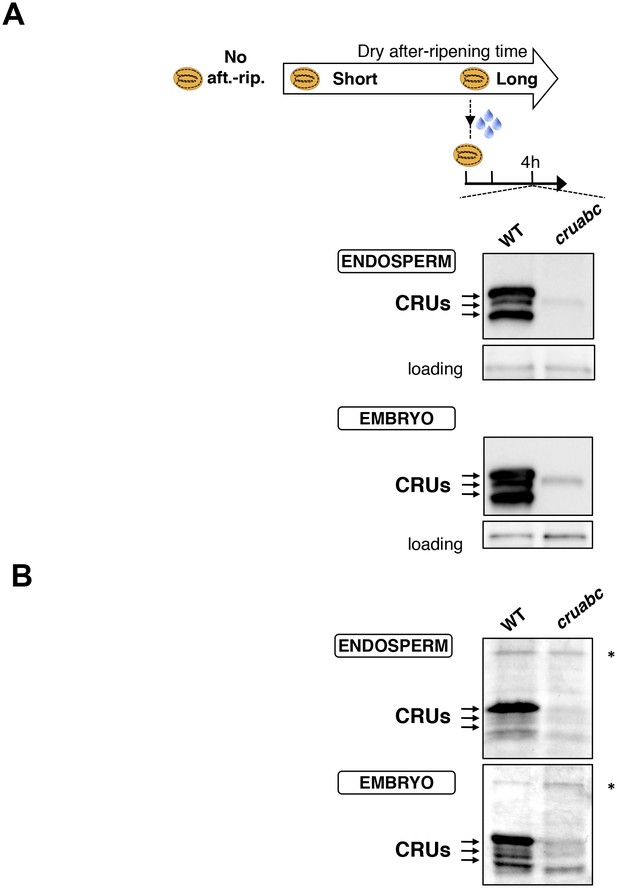
Antibody specificity.
(A) Protein gel blot analysis using 12S antibody serum. Protein extracts were isolated from the same number of WT (Col) and cruabc endosperms or embryos harvested 4 hr after seed imbibition. Proteins were separated on SDS PAGE gels. An aspecific band is used as a loading control. (B) Coomassie blue staining of proteins isolated from the same number of WT (Col) and cruabc endosperms or embryos separated on SDS PAGE gels. The asterisk (*) denotes a band used as an internal loading control.
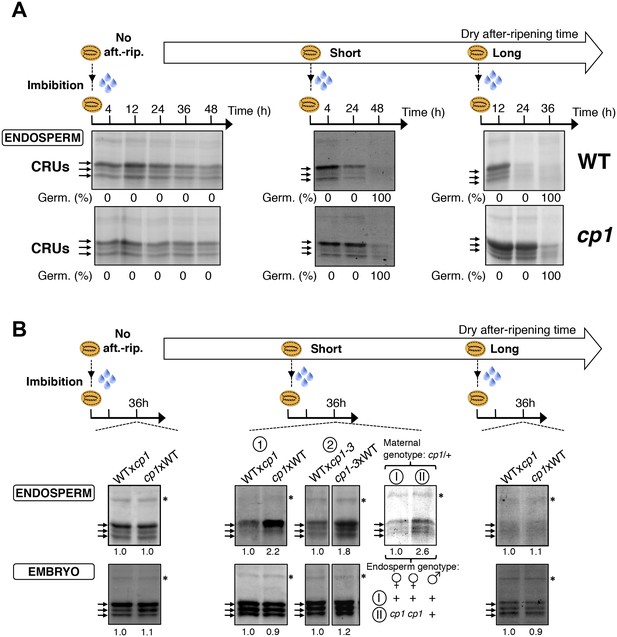
CP1 promotes CRU protein decay.
CRU protein decay is controlled through CP1 maternal gametophytic alleles. (A) Endosperms from non-after-ripened (No aft.-rip.) WT and cp1 seeds or from WT and cp1 seeds after-ripened for short (three days) or long (two months) time periods were dissected at the indicated times after seed imbibition. Proteins isolated from the same number of WT (Col) or cp1 endosperms were separated on SDS PAGE gels and stained with coomassie blue. For a given after-ripening time, WT and cp1 endosperm proteins were separated on the same SDS PAGE gel. An aspecific band is used as a loading control. Percent germination at each time-point after seed imbibition is indicated. Arrows indicate highly abundant cruciferin (CRU) proteins. (B) WTxcp1 F1 and cp1xWT F1 seeds (obtained by reciprocally crossing WT (Col) and cp1 plants) were subject to dry after-ripening treatments as in (A). For each after-ripening treatment, endosperms and embryos were dissected 36 hr after seed imbibition. Proteins isolated from the same number of endosperms or embryos were processed as in (A). Seeds obtained after crossing cp1/+ heterozygous mother plants with Col WT (+/+) pollen were after-ripened for a short time period (three days). Endosperms were dissected 36 hr after imbibition, whereas embryos were further cultured for later genotyping in order to distinguish endosperms according to their (i) +/+/+ or (ii) cp1/cp1/+ genotype. Endosperms with the same genotype were pooled. Proteins extracted from the two endosperms pools were processed as in (A). The asterisk (*) denotes a band used as an internal loading control. Numbers indicate protein signal quantification using imageJ software.
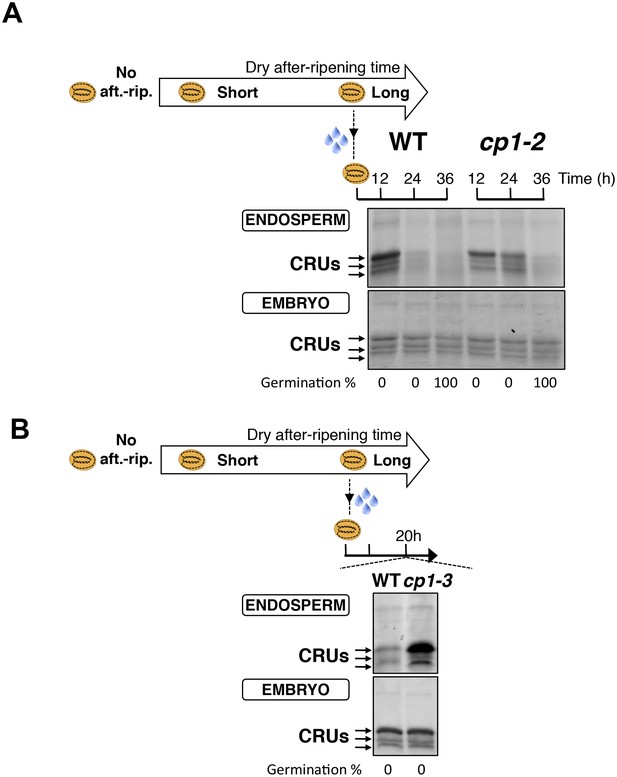
CP1 promotes CRU protein decay.
(A) Endosperms from fully after-ripened (two months) WT and cp1-2 (salk _020878) seeds were dissected at the indicated times after seed imbibition. Proteins isolated from the same number of WT (Col) or cp1-2 endosperms were separated on SDS PAGE gels and stained with coomassie blue. The WT samples are the same as those in Figure 4—figure supplement 2. Percent germination at each time-point after seed imbibition is indicated. Arrows indicate highly abundant cruciferin proteins (CRUs). (B) Endosperms from fully after-ripened (two months) WT and cp1-3 (salk _067293) seeds were dissected 20 hr after seed imbibition and processed as in (A).
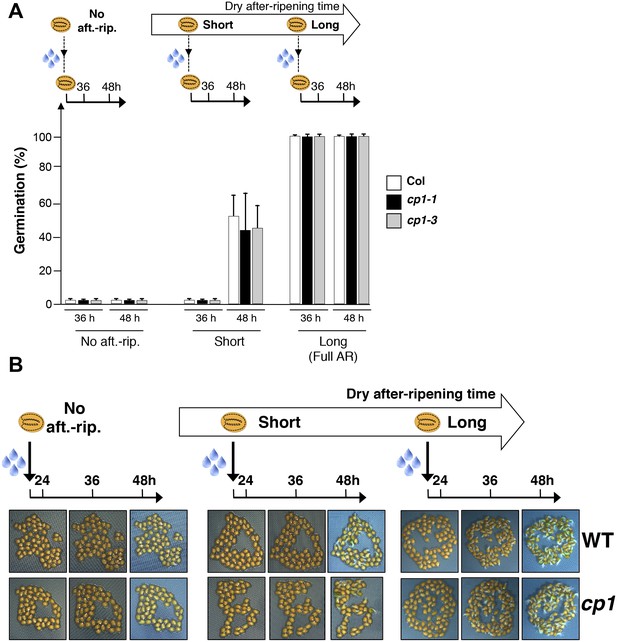
CP1 is not required to regulate germination sensu stricto.
(A) WT (Col), cp1-1 (salk_146500) and cp1-3 (salk_067293) seeds were either not after-ripened (No aft.-rip) or after-ripened for short (three days) or long (two months) time periods. Germination was scored 36 and 48 hr after seed imbibition (4 replicates, n = 150–200). (B) Representative pictures of WT of cp1-1 seeds 24, 36 and 48 hr after seed imbibition. After-ripening times as described in (A).
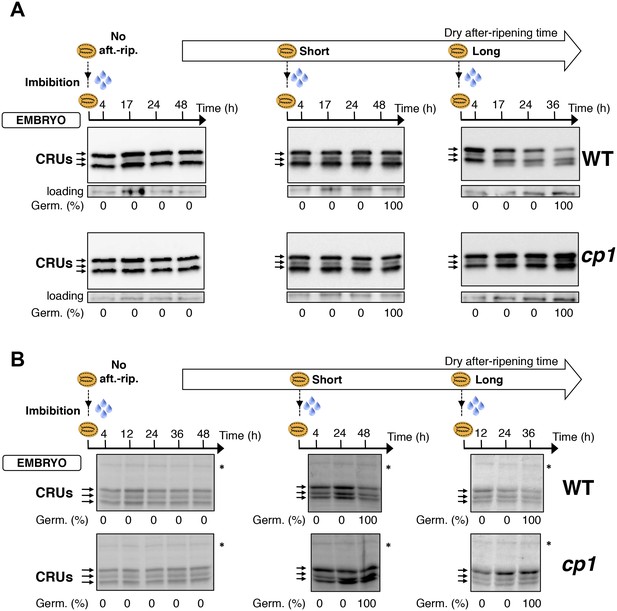
Embryo CRU protein levels are constant during early seed imbibition times.
Embryos from non-after-ripened (No aft.-rip.) WT (Col) and cp1 seeds or from WT and cp1 seeds after-ripened for short (three days) or long (two months) time periods were dissected at the indicated times after seed imbibition. Proteins isolated from the same number of embryos were separated on SDS PAGE gels and probed using 12S antibody serum (A) or stained with coomassie blue (B). For a given after-ripening time, WT and cp1 embryo proteins were separated on the same SDS PAGE gel. Percent germination at each time-point after seed imbibition is indicated. Arrows indicate highly abundant cruciferin proteins (CRUs). An aspecific band is used as a loading control in (A). The asterisk (*) denotes a band used as an internal loading control in (B).
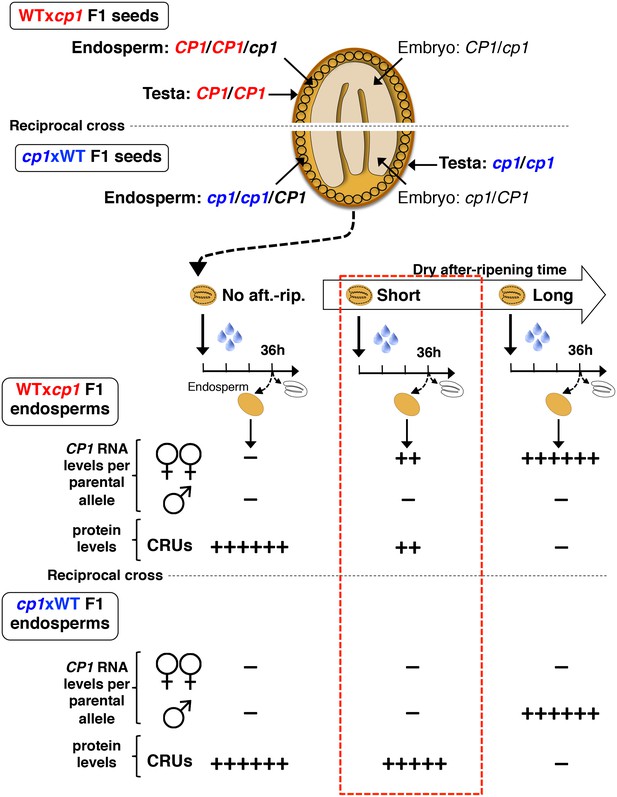
Procedure to assess CRUprotein decay through maternal CP1 alleles.
The genotypes of the different tissues present in WTxcp1 F1 and cp1xWT F1 seeds are depicted. In absence of after-ripening, CP1 maternal allele expression is not detectable, which leads to similarly high CRUs protein levels in the WTxcp1 F1 and cp1xWT F1 endosperms upon seed imbibition. In fully after-ripened seeds, CP1 expression is high and biallelic so that both WTxcp1 F1 and cp1xWT F1 endosperms are able to downregulate CRU proteins. By contrast, in shortly after-ripened seeds, preferential CP1 maternal allele expression takes place so that WTxcp1 F1 endosperm cells express higher CP1 mRNA levels than do cp1xWT F1 endosperms. As a result, upon imbibition, WTxcp1 F1 endosperms ought to accumulate lower CRUlevels relative to cp1xWT F1 endosperms.
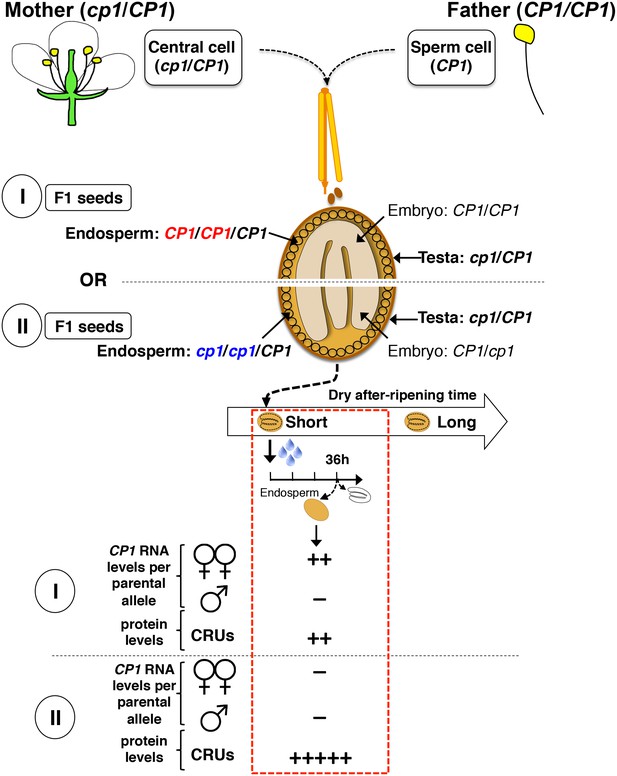
Procedure for the assessment of CRU protein decay through CP1 maternal gametophytic alleles.
Hybrid seeds were obtained after crossing heterozygous cp1/Col mother plants with WT (Col) pollen. The resulting siliques have two types of seeds according to their endosperm tissues: type I bear CP1 maternal genomes, whereas type II bear cp1 maternal genomes. Shortly after-ripened type I seeds are expected to have endosperm tissues that express higher CP1 mRNA levels than do type II seeds. Thus, endosperm from type I seeds should have lower CRU levels relative to that of type II seeds upon seed imbibition.
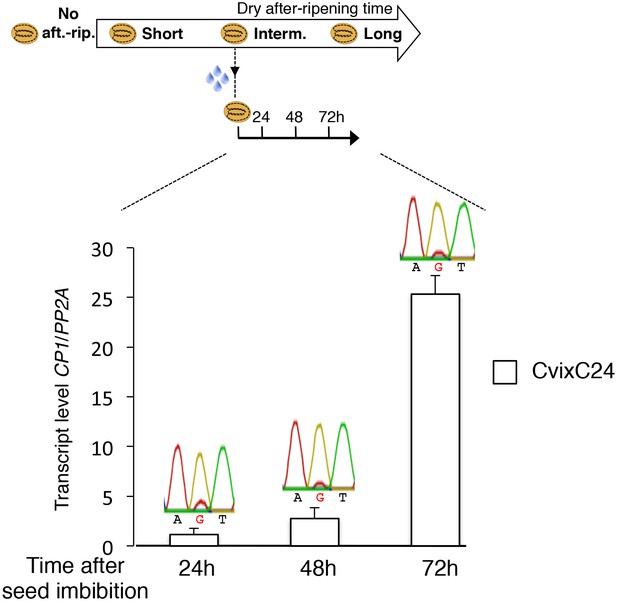
Monoallelic CP1 expression rises upon imbibition in dormant seeds.
CvixC24 (C24 pollen) F1 seeds were after-ripened for an intermediate time period (25 days). RNA extracted from endosperm 24 hr, 48 hr and 72 hr after seed imbibition, i.e. prior to germination, was used for qPCR and RT-PCR for Sanger sequencing analysis. Expression levels of CP1 were normalized to those of PP2A. CP1 expression levels relative to those found in endosperms dissected at indicated time are shown.
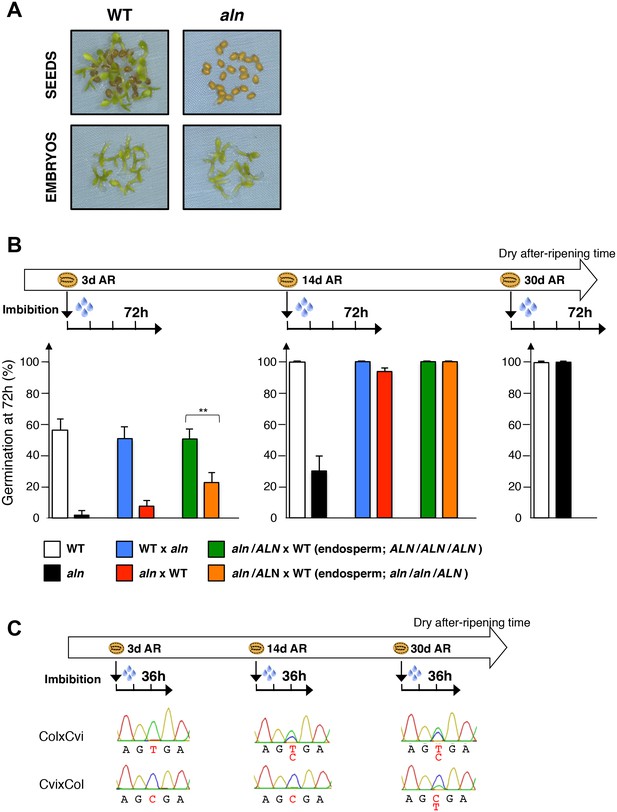
ALN affects seed dormancy level through maternal alleles.
(A) WT and aln mutant seeds were after-ripened for 10 days. Pictures show seeds (upper panel) and embryos (lower panel) 72 hr after imbibition. Embryos were dissected 4 hr after imbibition. (B) WT, aln, WTx aln F1, aln x WT F1 and aln/ALN x WT F1 seeds were after-ripened for 3 days, 14 days, and 30 days (only for WT and aln seeds). Germination was scored 72 hr after seed imbibition (4 replicates, n = 50, **p<0.01, Student’s t test). After germination, aln/ALN x WT individuals were genotyped to distinguish endosperms bearing aln/aln/ALN and ALN/ALN/ALN alleles. (C) Sanger sequencing chromatograms covering SNPs present in ALN. CvixCol and ColxCvi F1 seeds were dry after-ripened for 3 days, 14 days, and 30 days. RNA extracted from endosperm dissected 36 hr after imbibition was processed for RT-PCR followed by Sanger sequencing. Nucleotides at SNP sites are highlighted with red.
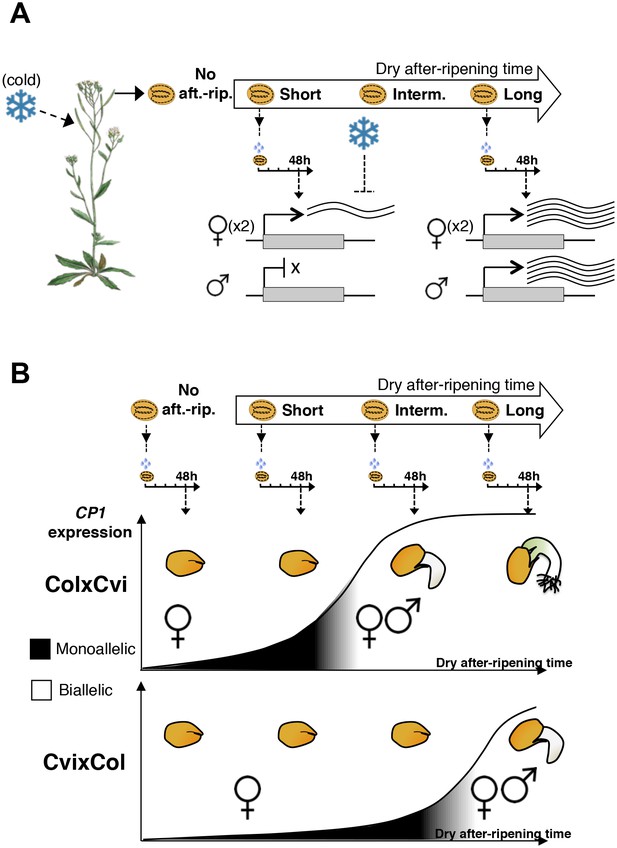
A model for maternal dormancy inheritance in seed hybrids.
(A) General mechanism accounting for maternal inheritance of seed dormancy in hybrid seeds. Upon imbibition, a group of genes that regulate seed germination processes are preferentially maternally expressed during the seed-dormant period. As a result, dormancy levels in hybrid seeds reflect the maternal genotype. (B) The case of CP1. In the absence of after-ripening (No aft.-rip), preferential maternal allele expression is very low. As seeds begin to after-ripen (Short), preferential maternal allele expression rises according to the maternal genotype, i.e. there is higher maternal allelic expression in ColxCvi F1 relative to CvixCol F1 seeds (see also Figure 3). After an intermediate after-ripening period, low and maternal allelic CP1 expression is lost in ColxCvi F1 seeds but not in CvixCol F1 seeds. As a result, seed CRUs decay is initiated earlier in ColxCvi F1 seeds relative to CvixCol F1 seeds. Further after-ripening abolishes preferential maternal allelic expression in CvixCol F1 seeds upon imbibition, which leads to undelayed decay of CRU proteins and seed germination.
Tables
RNA concentrations and read number after RNA-seq using RNA isolated from partially dissected endosperm (with testa still attached to the endosperm), fully dissected endosperm, and testa.
Sample | RNA concentration ng/µl in 50 µl volume | Total amount of RNA (ng) | Volume of RNA sample used for library preparation (µl) | cDNA library (ng/µl) | Volume of library used for sequencing | Total mapped sequencing reads |
|---|---|---|---|---|---|---|
Partially dissected endosperm_A | 5 | 250 | 40 | 37.4 | 1.16 | 39,097,682 |
Partially dissected endosperm_B | 6.4 | 320 | 31.25 | 15.2 | 2.5 | 31,930,392 |
Fully dissected endosperm_A | 7 | 350 | 28.57 | 16.4 | 2.5 | 39,862,912 |
Fully dissected endosperm_B | 4.4 | 220 | 45.45 | 15.8 | 2.5 | 35,254,459 |
Testa_A | 0 | 0 | 45.45 | 0.37 | 2.5 | 264,057 |
Testa_B | 0 | 0 | 45.45 | 0.59 | 2.5 | 595,710 |
Read numbers of the identified MEGs in testa, partially dissected endosperm (endosperm + testa), and fully dissected endosperm samples.
Testa | Partially dissected endosperm | Fully dissected endosperm | |
|---|---|---|---|
AT4G00220 | 9 | 603 | 597 |
AT4G04955 | 3 | 2,154 | 1,859 |
Additional files
-
Supplementary file 1
List of germination-related MEGs.
- https://doi.org/10.7554/eLife.19573.035
-
Supplementary file 2
Primers used in this study.
- https://doi.org/10.7554/eLife.19573.036
-
Supplementary file 3
Cvi SNPs data.
- https://doi.org/10.7554/eLife.19573.037





