Pathogenic PS1 phosphorylation at Ser367
Figures
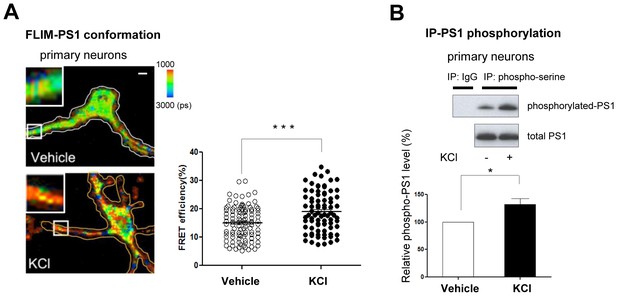
Ca2+ influx-triggered PS1 conformational change and increased phosphorylation.
(A) FLIM analysis of PS1 conformation. Pseudo-colored FLIM images of primary neurons treated with vehicle control or 50 mM KCl for 5 min. The neurons were stained with the antibodies to PS1 NT (Alexa488) and PS1 CT (Cy3). The colorimetric scale shows Alexa488 lifetime in picoseconds (ps). A scale bar indicates 10 µm. The graph shows quantitative analysis of the FRET efficiency between fluorescently labeled PS1 NT and PS1 CT in neuronal processes (total of 81–103 processes from 32–38 cells). Mean ± SEM, ***p<0.001, Student’s t-test. (B) Immunoprecipitation/Western blot analysis of PS1 phosphorylation. Primary neurons treated with vehicle control or 50 mM KCl for 5 min were immunoprecipitated with mouse and rabbit anti-phosphoserine antibody mixture, followed by immunoblotting with the anti-PS1 loop antibody (Upper panel, top row). Normal mouse and rabbit IgG mixture was used as negative control. Lower gel shows the total level of PS1 CTF in neuronal cell lysates. The graph presents a quantitative analysis of the band intensity for phosphorylated PS1 (n = 6). The relative PS1 phosphorylation level in KCl-treated neurons was normalized to that in vehicle-treated cells. Mean ± SEM, *p<0.05, One sample t-test.
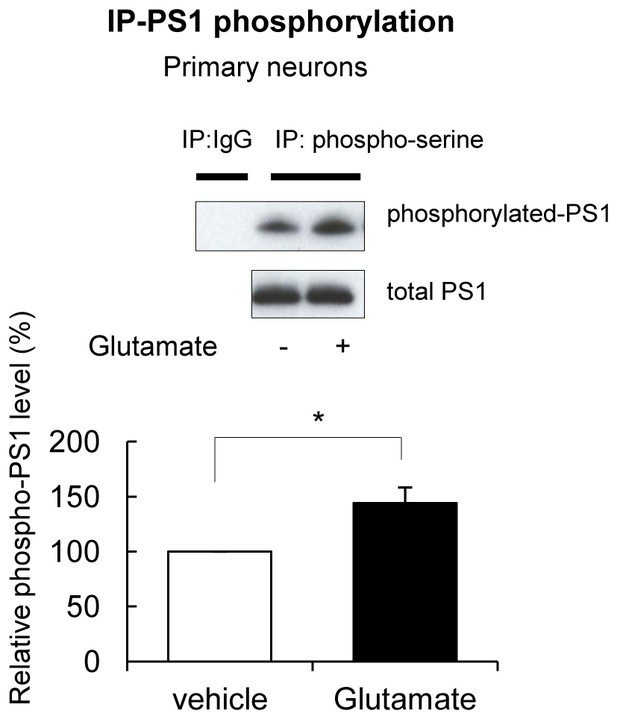
Glutamate treatment induces PS1 phosphorylation.
Primary neurons treated with 5 mM glutamate or with vehicle control for 2 min, were immunoprecipitated with mouse and rabbit anti-phosphoserine antibodies mixture, followed by the immunoblotting with an anti-PS1 loop antibody. Glutamate does not affect total PS1 CTF levels. The graph shows the relative PS1 phosphorylation level in glutamate-treated neurons normalized to that in vehicle-treated neurons (n = 6). Mean ± SEM, *p<0.05, One sample t-test.
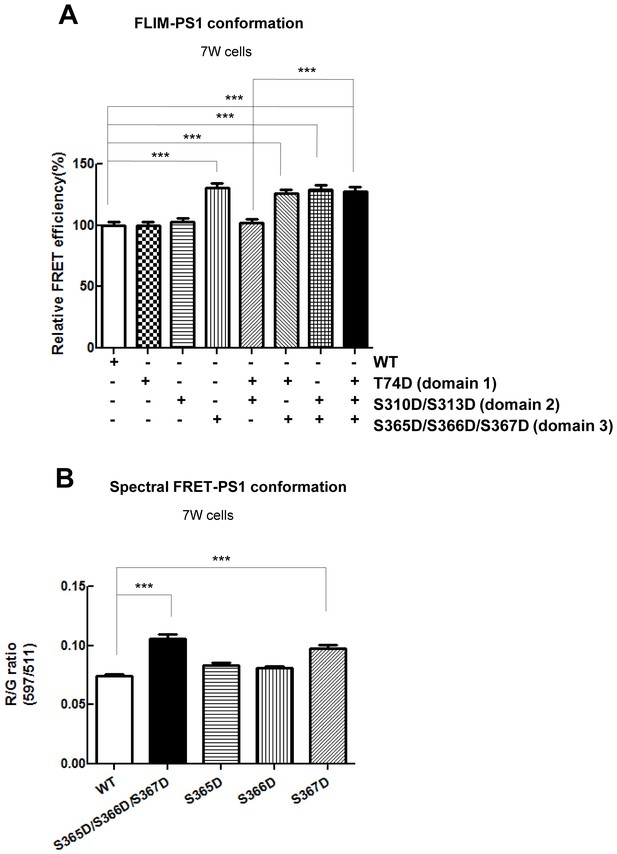
Phosphorylation mimicking mutations at S365-S366-S367 result in the PS1 conformational shift.
(A) FLIM analysis of the PS1 conformation in 7 W cells transfected with WT or phosphorylation-mimicking mutant G-PS1-R. The FRET efficiency between GFP and RFP in phospho-mutants is normalized to the average FRET efficiency of the WT G-PS1-R expressing cells (n = 42–53 cells). Mean ± SEM, ***p<0.001, one-way factorial ANOVA. (B) Spectral FRET analysis shows RFP/GFP (R/G) ratio in 7 W cells transfected with WT or single phosphorylation-mimicking mutant G-PS1-R (n = 50–69 cells). Mean ± SEM, ***p<0.001, one-way factorial ANOVA.
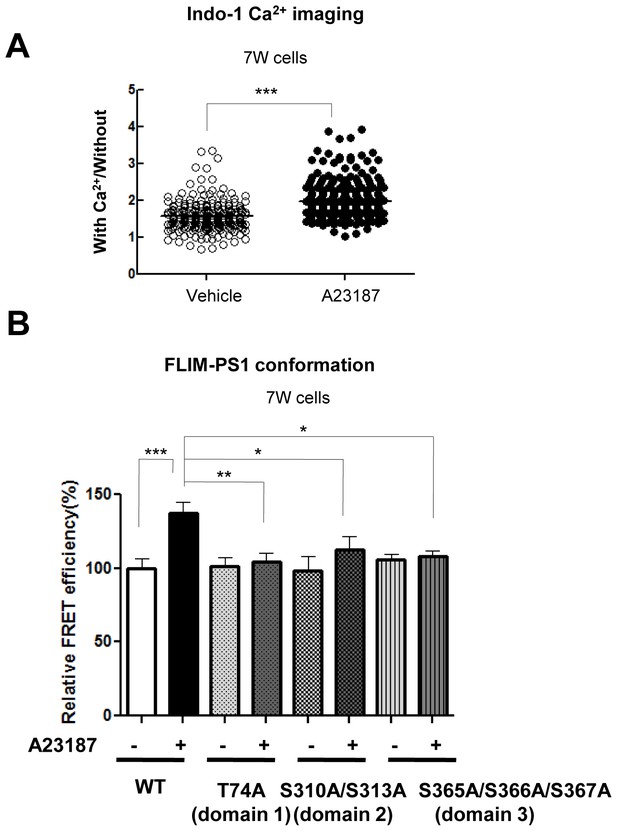
Calcium imaging and validation of the phosphorylation at the three domains-driven PS1 conformational change by the G-PS1-R FRET reporter probe.
This figure is related to the data displayed in Table 1. (A) Indo-1/Ca2+ imaging shows changes in intracellular Ca2+ levels of the 7 W cells transfected with FLAG-WT PS1, following 15 min treatment with vehicle control or with 5 µM A23187 (n = 250–297 cells). Mean ± SEM, ***p<0.001, Student’s t-test. (B) FLIM analysis of the PS1 conformation in the 7 W cells transfected with WT or domain 1, 2 or 3 phosphorylation-inhibited mutants PS1, following 15 min treatment with vehicle control or with 5 µM A23187. The FRET efficiency in each group is normalized to the average FRET efficiency of vehicle-treated cells expressing WT G-PS1-R (n = 22–55 cells). Mean ± SEM, *p<0.05, **p<0.01, ***p<0.001, one-way factorial ANOVA.
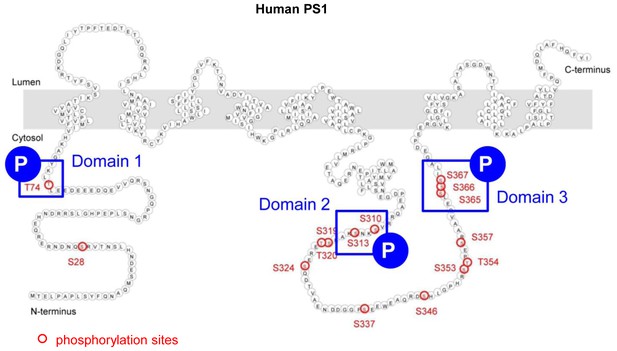
Schematic image of the three domains in PS1 involved in Ca2+-triggered pathogenic ‘closed’ conformation.
This figure is related to the data displayed in Table 1. Schematic image of PS1 molecule and the three domains involved in Ca2+-triggered PS1 pathogenic ‘closed’ conformation (blue squares). Serine/threonine residues known to undergo phosphorylation are shown by red circles.
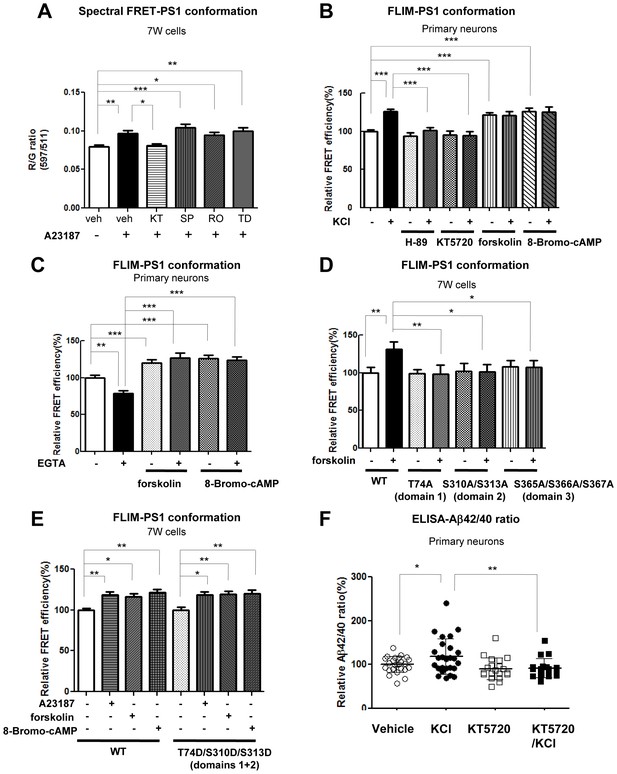
PKA activity is involved in the Ca2+-driven PS1 conformational change.
(A) Spectral FRET analysis of the PS1 conformation in 7 W cells transfected with WT G-PS1-R and pre-treated with 1 µM KT5720 (KT, PKA inhibitor, 16 hr), 20 µM SP600125 (SP, JNK inhibitor, 4 hr), 100 nM Ro 31–8220 (Ro, PKC inhibitor, 16 hr), or 5 µM TDZD-8 (TD, GSK3β inhibitor, 4 hr). Spectral FRET imaging was performed after the cells were treated with DMSO (vehicle control) or 5 µM A23187 for 15 min (n = 31–50 cells). Mean ± SEM, *p<0.05, **p<0.01, ***p<0.001, one-way factorial ANOVA. (B) Endogenous PS1 conformation was monitored by the antibody-based FLIM analysis. Primary neurons were pre-treated with PKA inhibitors: 30 µM H-89 or 1 µM KT5720 for 16 hr, or with PKA activators: 10 µM forskolin or 0.5 mM 8-Bromo-cAMP for 1 hr, followed by the treatment with vehicle control (-) or 50 mM KCl (+) for 5 min. The FRET efficiency in each group is normalized to the average FRET efficiency of vehicle control treated neurons (n = 36–125 processes from n = 17–28 cells). Mean ± SEM, ***p<0.001, one-way factorial ANOVA. (C) Ca2+ sequestration by EGTA affects PS1 conformation in vehicle but not PKA activator-treated neurons. Primary neurons were pre-treated with vehicle, 10 µM forskolin or 0.5 mM 8-Bromo-cAMP for 1 hr, followed by the treatment with 2 mM EGTA or vehicle control for 15 min. The FRET efficiency in each group is normalized to the average FRET efficiency of vehicle control neurons (n = 19–34 cells, n = 40–103 processes). Mean ± SEM, **p<0.01, ***p<0.001, one-way factorial ANOVA. (D) FLIM analysis of the PS1 conformation in 7 W cells transfected with WT or phosphorylation-inhibited mutants G-PS1-R, and treated with 10 µM forskolin or vehicle control for 1 hr. The FRET efficiency is normalized to the average FRET efficiency of vehicle-treated cells expressing WT G-PS1-R (n = 20–39 cells). Mean ± SEM, *p<0.05, **p<0.01, one-way factorial ANOVA. (E) FLIM analysis of the PS1 conformation in 7 W cells transfected with WT or domain 1 + 2 phosphorylation-mimicking G-PS1-R isoform, and treated with 5 µM A23187, 10 µM forskolin or 0.5 mM 8-Bromo-cAMP. The FRET efficiency normalized to that in vehicle treated cells is shown (n = 34–47 cells). Mean ± SEM, *p<0.05, **p<0.01, one-way factorial ANOVA. (F) KCl-induced increase of the Aβ42/40 ratio is prevented by PKA inhibitor. Primary neurons were pre-treated with vehicle control or 1 µM KT5720 for 16 hr, followed by the treatment with vehicle control or 50 mM KCl for 30 min. Aβ40 and Aβ42 levels were measured by ELISA. The Aβ42/40 ratio in each group is normalized to that of the vehicle control neurons (n = 19–25). Mean ± SEM, *p<0.05, **p<0.01, one-way factorial ANOVA.
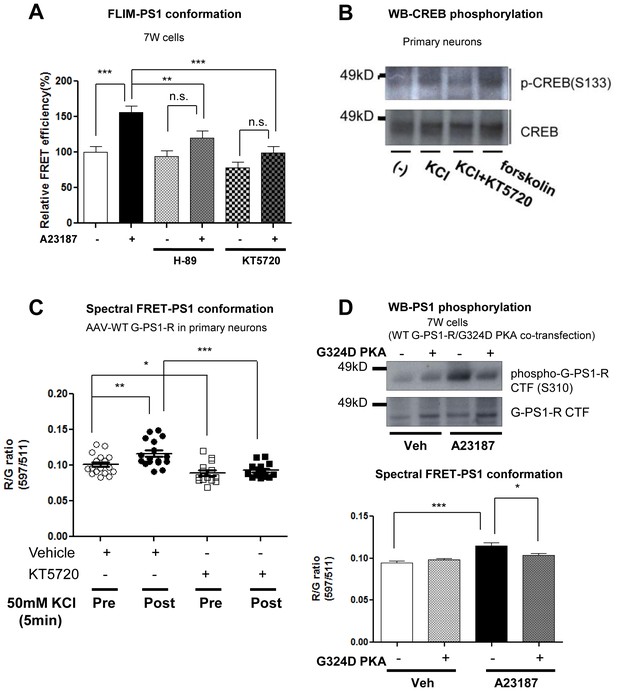
PKA is involved in the Ca2+-triggered PS1 pathogenic ‘closed’ conformation.
(A) FLIM analysis of the PS1 conformation in 7 W cells. The cells were pre-treated with 30 µM H-89 or 1 µM KT5720 for 16 hr, followed by the treatment with vehicle control (−) or 5 µM A23187 (+) for 15 min. The FRET efficiency in each group is normalized to the average FRET efficiency of vehicle control treated neurons (n = 37–52 cells). Mean ± SEM, **p<0.01, ***p<0.001, one-way factorial ANOVA. (B) Western Blot analysis of PKA-dependent CREB S133 phosphorylation. Primary neurons were pre-treated with vehicle control or 1 µM KT5720 for 16 hr, followed by the treatment with vehicle control or 50 mM KCl for 5 min. Treatment with 10 µM forskolin was used as a positive control of PKA activation. (C) Spectral FRET assay of the PS1 conformation in primary neurons. AAV-G-PS1-R-transduced primary neurons were pre-treated with 1 µM KT5720 for 16 hr, followed by the treatment with 5 mM KCl for 5 min. Pre: before KCl application, Post: 5 min after KCl application. n = 14–17 cells, Mean ± SEM, *p<0.05, **p<0.01, ***p<0.001, one-way factorial ANOVA. (D) Dominant negative PKA regulatory subunit α (G324D PKA) was co-transfected with WT G-PS1-R into 7 W cells. The cells were treated with vehicle control or 5 µM A23187 for 15 min. Western Blot analysis of the PS1 S310 phosphorylation (top panel). Spectral FRET assay of the PS1 conformation (bottom panel). n = 83–88 cells, Mean ± SEM, *p<0.05, ***p<0.001, one-way factorial ANOVA.
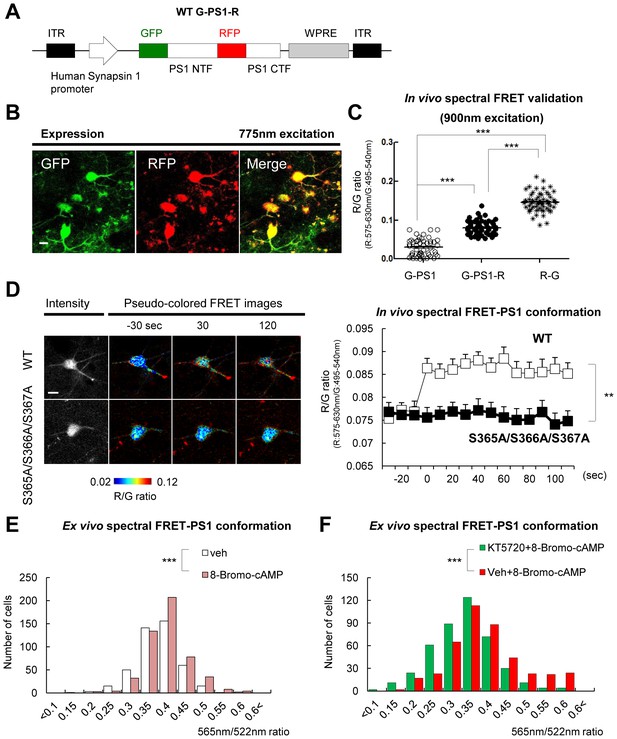
Ca2+ triggers the PS1/γ-secretase pathogenic conformation via PS1 phosphorylation in vivo.
(A) Schematic representation of the pAAV8-hSyn1-WT G-PS1-R construct. (B) Two-photon image of the WT G-PS1-R expression in the somatosensory cortex of WT mouse. Laser at 775 nm wavelength was used for the excitation. A scale bar indicates 10 µm. (C) Mice were injected with AAV8-hSyn1-G-PS1 (as a negative control of FRET), AAV8-hSyn1-WT G-PS1-R or AAV8-hSyn1-R-G (as a positive control of FRET). GFP was excited at 900 nm wavelength, and the R/G ratio was recorded (n = 50–60 cells, n = 3–6 mice). Mean ± SEM, ***p<0.001, one-way factorial ANOVA. (D) Spectral FRET analysis of the PS1 conformation in vivo. Mice were injected with AAV8-hSyn1-WT G-PS1-R (n = 3) or AAV8-hSyn1-S365A/S366A/S367A G-PS1-R (n = 4), and 300 mM KCl was applied topically. The R/G ratio in vivo was monitored after two-photon excitation at 900 nm (total n = 20–28 cells per condition) for the duration of 2 min. Representative images of the pseudo-colored neurons are shown (additional time traces/images are shown in Figure 4—figure supplement 3). Mean ± SEM, **p<0.01, two-way repeated-measures ANOVA. (E) Ex-vivo spectral FRET analysis of the endogenous PS1 conformation in mouse brain sections. Mice were injected with 100 mM 8-Bromo-cAMP (right hemisphere) or vehicle (left hemisphere) into the somatosensory cortex. The 565 nm/522 nm ratio was calculated in individual neurons as readout of the FRET efficiency that reflects the relative proximity of the PS1 NT (A488) to PS1 CT (Cy3). The histogram shows cell numbers plotted against the 565 nm/522 nm ratios. n = 3 mice, total of 444 (vehicle) and 505 (8 Bromo-cAMP) neurons. ***p<0.001, Student’s t-test. (F) Ex-vivo spectral FRET analysis of the endogenous PS1 conformation in mouse brain sections. Mice were pre-injected with 100 µM KT5720 (right hemisphere) or vehicle (left hemisphere) into the somatosensory cortex. 75 min post-injection, 100 mM 8-Bromo-cAMP (both hemispheres) was delivered to the same area for 5 min. The histogram shows cell numbers plotted against the 565 nm/522 nm ratios. n = 3 mice, total 423 (vehicle) and 436 (KT5720) neurons analysed. ***p<0.001, Student’s t-test.
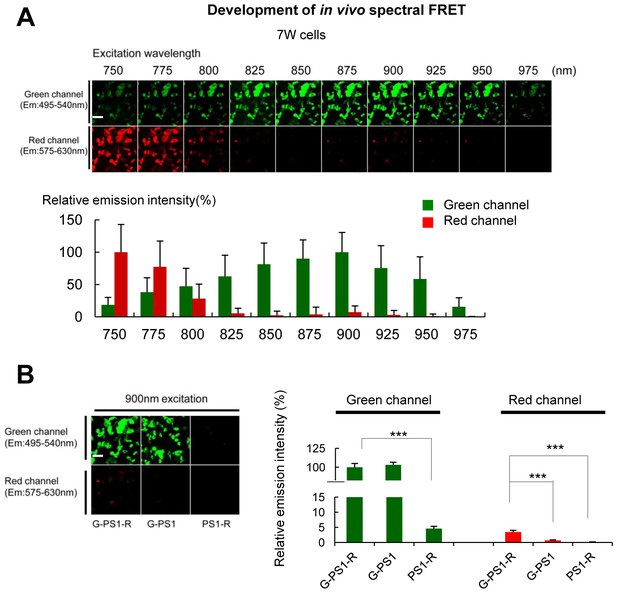
Establishment of the two-photon spectral FRET settings for monitoring PS1 conformation.
(A) 7 W cells expressing G-PS1-R were excited by different wavelength laser from 750 nm to 975 nm in 25 nm steps. The top panel shows representative images of the emission intensities in green and red channel (n = 41). A scale bar indicates 20 µm. The graph shows quantitative analysis of the relative emission intensities, with the intensity at 900 nm (green channel) and 750 nm (red) as 100%. Mean ± SEM. (B) 7 W cells expressing G-PS1-R, G-PS1 or PS1-R were excited by 900 nm laser. Left panel shows the representative images of relative emission intensities in green or red channel. A scale bar indicates 20 µm. The intensity of G-PS1-R in green channel was set as 100% (n = 36 each). Mean ± SEM, ***p<0.001, one-way factorial ANOVA.
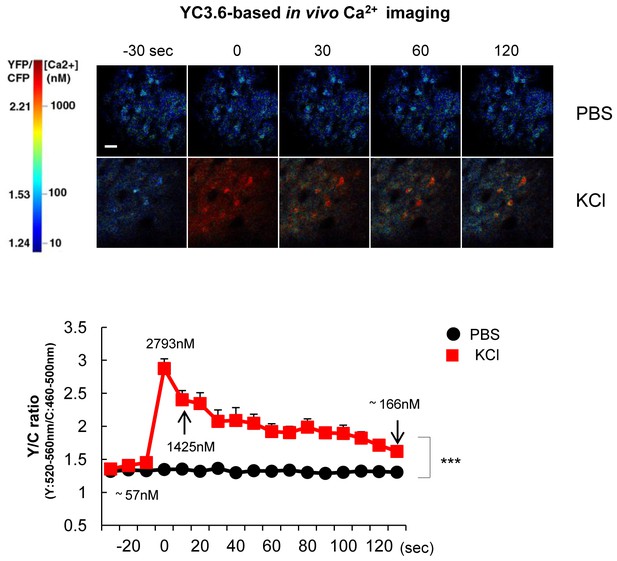
YC3.6-based Ca2+ imaging in vivo after KCl application.
300 mM KCl or vehicle (PBS) was topically applied under the cranial window to mice expressing YC3.6. The YFP/CFP ratio was measured in neurons of the somatosensory cortex in vivo (n = 18–22 cells, three different mice for each group). Representative pseudo-colored images were shown in the top panels. Mean ± SEM, ***p<0.001, two-way repeated-measures ANOVA.
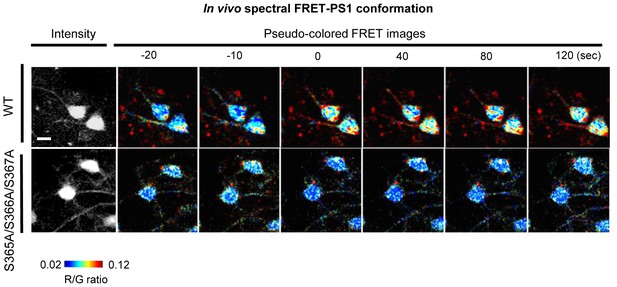
Spectral FRRT assay of PS1 conformation in vivo.
Additional time traces and pseudo-coloured images of the neurons in vivo showing changes in PS1 conformation. A scale bar indicates 10 µm.
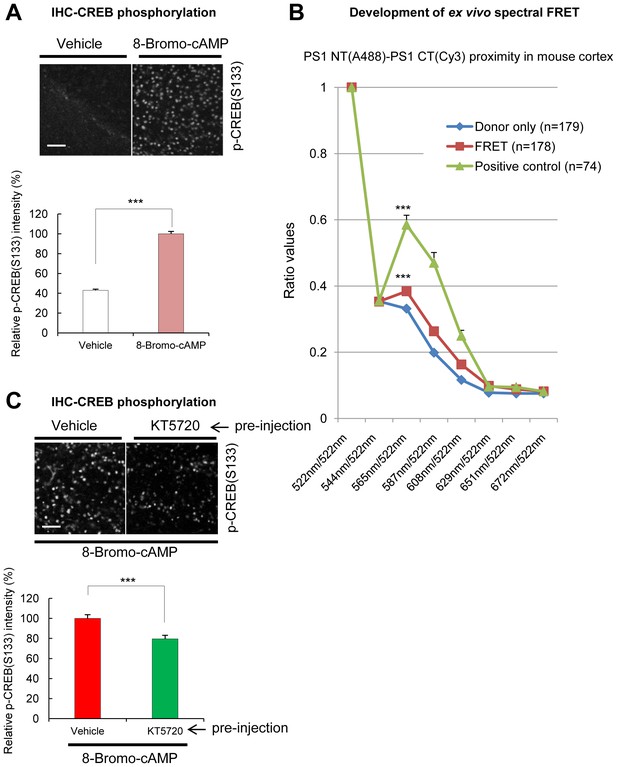
Spectral FRET assay for monitoring endogenous PS1 conformation in mouse brain sections.
(A) Immunohistochemical detection of the phosphorylated CREB in mouse brain injected with 100 mM 8-Bromo-cAMP (right hemisphere) or vehicle (left hemisphere). The brain sections were immunostained with a p-CREB S133 antibody. Strong fluorescence in cAMP injected hemisphere shows PKA activation. A scale bar indicates 50 µm. Mean ± SEM, ***p<0.001, Student’s t-test. (B) Antibody-based spectral FRET validation. The brain sections were immunostained with PS1 NT-A488 (donor only, negative control for FRET), PS1 NT-A488-Cy3 (A488-Goat-anti-mouse IgG was detected with Cy3-Donkey-anti-goat IgG, positive control), or PS1 NT-A488/PS1 CT-Cy3 (FRET, PS1 NT-PS1 CT proximity) antibodies. The Y axis shows the ratio of fluorescence intensities within the 522 nm to 672 nm spectral range normalized by the emission intensity at 522 nm. The positive control (in green) shows large ‘hump’ at 565/522 nm, and the FRET signal in PS1 NT-PS1 CT stained cells (in red) shows smaller but significant increase in the 565 nm/522 nm ratio compared to the donor only (n = 75–179 cells). Mean ± SEM, ***p<0.001 vs. Donor only, one-way factorial ANOVA. (C) Immunohistochemical detection of the phosphorylated CREB S133 in mouse brain injected with 100 µM KT5720 (right hemisphere) or vehicle (left hemisphere), followed by the injection of 100 mM 8-Bromo-cAMP (both hemispheres). A scale bar indicates 50 µm. Mean ± SEM, ***p<0.001, Student’s t-test.
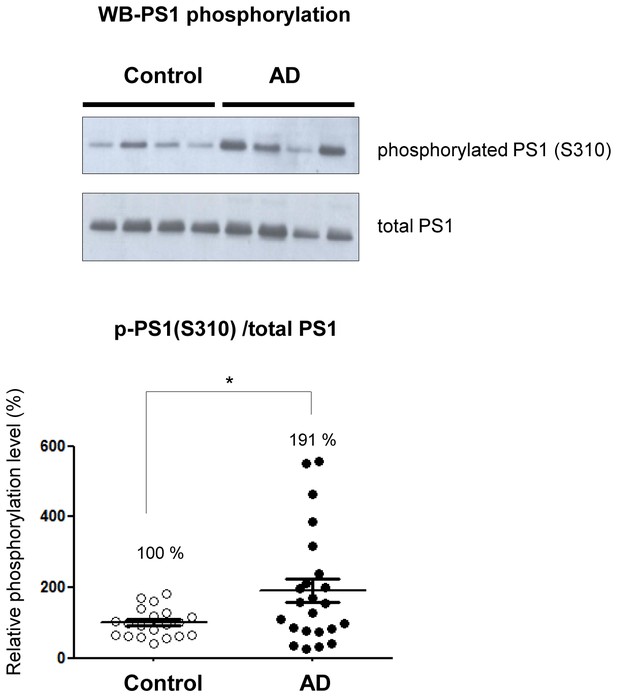
Phosphorylated PS1 level is increased in AD brains.
Western blot analysis of PS1 phosphorylated at S310 in 20 control brains and 23 AD brains. The level of PS1 S310 phosphorylation (phospho-PS1/total PS1) in AD brains is normalized to that in the control brains. Mean ± SEM, *p<0.05, Student’s t-test.
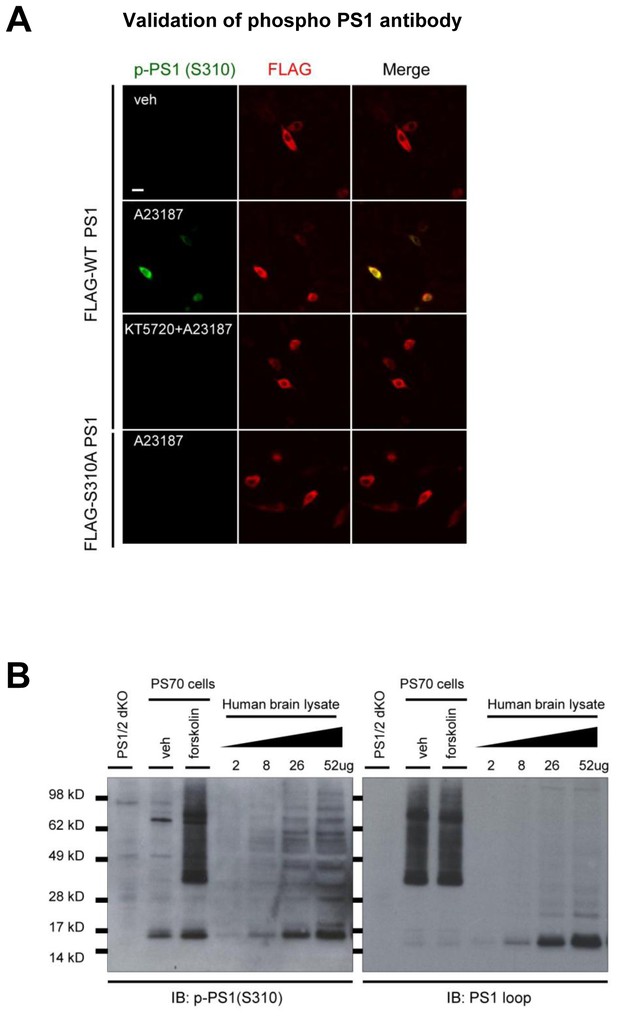
Validation of the PKA-mediated PS1 phosphorylation at S310.
(A) Confocal microscope images of the 7 W cells immunostained with anti-phosphorylated PS1 at S310 (green) or anti-FLAG antibodies (red). 7 W cells transfected with FLAG-WT PS1 or FLAG-S310A PS1 were pre-treated with 1 µM KT5720 or vehicle control for 16 hr, followed by the treatment of 5 µM A23187 or vehicle control for 15 min. A scale bar indicates 20 µm. (B) The level of phosphorylated PS1 at S310 was compared between forskolin and vehicle-treated PS70 cells by Western blotting. PS1/2 dKO MEF cells were used as a negative control. The level of phosphorylated PS1 at S310 was measured in the AD brain sample loaded at different total protein concentration.
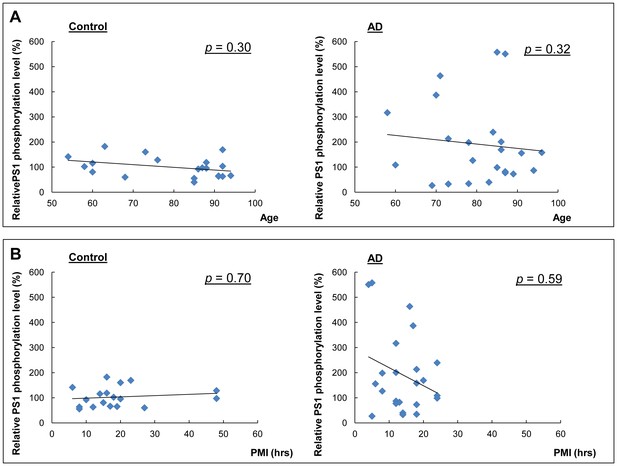
Correlation between the relative phosphorylation level of PS1 and age, or PMI.
Correlation analysis of phosphorylation of PS1 at S310 with age in control and AD brains (A), or with post mortem interval (PMI) (B). Spearman's nonparametric correlation analysis.
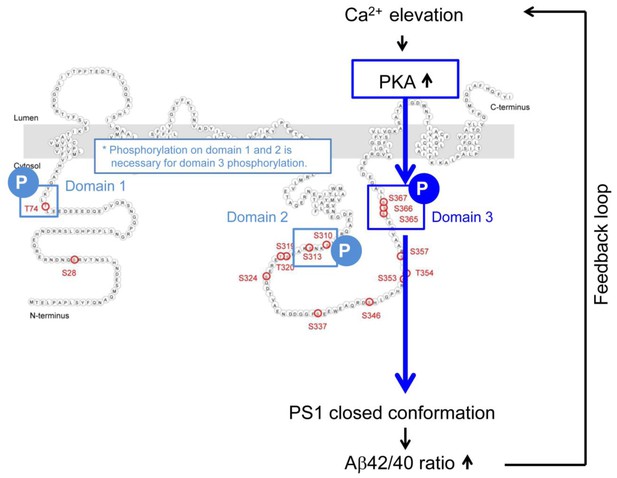
Mechanism of the Ca2+-triggered PS1 pathogenic conformational change.
The schematic image of the molecular events involved in the Ca2+-triggered pathogenic ‘closed’ conformational change and increase of the Aβ42/40 ratio. The elevated Ca2+ levels induce PKA activation, followed by the phosphorylation of PS1 at domain 1, domain 2 and domain 3. Domain 3 phosphorylation, particularly at S367, induces the PS1 pathogenic conformation that leads to increase in the Aβ42/40 ratio.
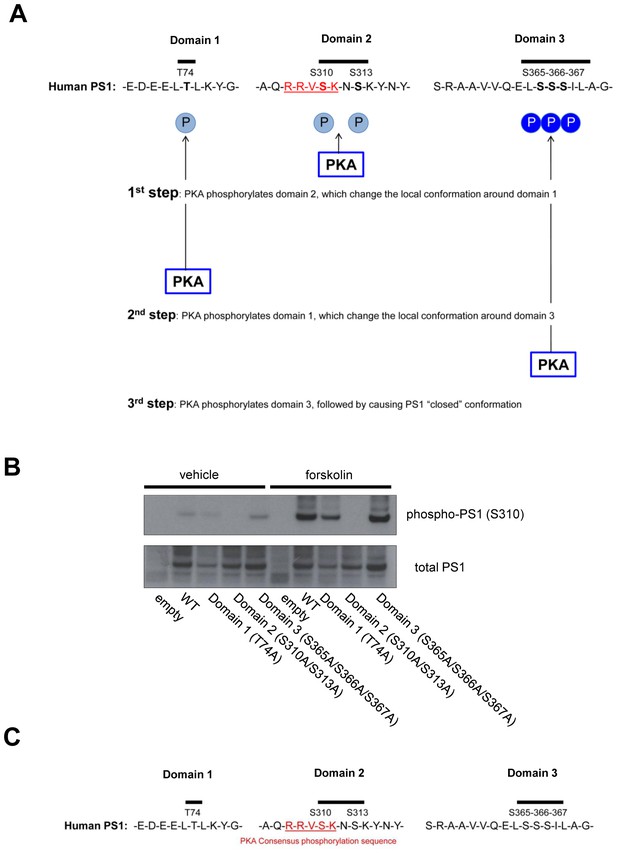
Model of the PKA-mediated PS1 phosphorylation.
(A) Model. PKA first phosphorylates domain 2, followed by changing local conformation around domain 1. PKA then phosphorylates domain 1, which subsequently leads to local rearrangement around domain 3. Finally, PKA phosphorylates domain 3, causing PS1 pathogenic conformation. (B) The level of phosphorylated PS1 at S310 was compared between forskolin and vehicle-treated 7 W cells expressing WT, domain 1, 2 or 3 phospho-inhibited PS1 by Western blotting. (C) Amino acid sequences around domain 1, 2 and 3. Domain 2 includes the PKA substrate consensus sequence: R-R-V-S-K.
Tables
FLIM analysis of the PS1 NT-CT proximity in phosphorylation-inhibited PS1 mutants.
Construct | *Relative FRET efficiency (%) | p value | ||
|---|---|---|---|---|
DMSO | A23187 | ‡vs WT (in DMSO) | §DMSO vs A23187 | |
PS1 Wild type (WT) | 100 ± 8.9 (n = 18) | 134.7 ± 7.5 (n = 25) | - | †p<0.05 |
PS1 S28A | 106.9 ± 7.0 (n = 19) | 143.4 ± 8.6 (n = 19) | n.s. | †p<0.05 |
PS1 T74A | 96.5 ± 13.9 (n = 14) | 98.5 ± 8.3 (n = 22) | n.s. | n.s. |
PS1 S310A | 88.8 ± 10.2 (n = 17) | 95.1 ± 8.6 (n = 24) | n.s. | n.s. |
PS1 S313A | 92.7 ± 8.4 (n = 15) | 108.0 ± 8.8 (n = 21) | n.s. | n.s. |
PS1 S310A/S313A | 84.2 ± 6.7 (n = 26) | 87.8 ± 9.0 (n = 17) | n.s. | n.s. |
PS1 S319A/T320A | 106.2 ± 8.1 (n = 20) | 131.8 ± 8.1 (n = 20) | n.s. | †p<0.05 |
PS1 S324A | 93.8 ± 6.2 (n = 19) | 129.9 ± 8.4 (n = 15) | n.s. | †p<0.05 |
PS1 S337A | 99.9 ± 8.8 (n = 13) | 132.4 ± 10.9 (n = 16) | n.s. | †p<0.05 |
PS1 S346A | 100.1 ± 8.9 (n = 18) | 135.0 ± 5.2 (n = 19) | n.s. | †p<0.05 |
PS1 S353A | 89.6 ± 7.4 (n = 18) | 129.8 ± 8.3 (n = 23) | n.s. | †p<0.05 |
PS1 T354A | 74.4 ± 5.6 (n = 14) | 113.4 ± 7.7 (n = 23) | n.s. | †p<0.05 |
PS1 S357A | 91.2 ± 7.9 (n = 18) | 119.8 ± 9.2 (n = 23) | n.s. | †p<0.05 |
PS1 S353A/S357A | 98.9 ± 9.1 (n = 14) | 125.9 ± 7.1 (n = 13) | n.s. | †p<0.05 |
PS1 S365A | 102.3 ± 6.4 (n = 19) | 87.3 ± 9.9 (n = 21) | n.s. | n.s. |
PS1 S366A | 102.8 ± 6.7 (n = 16) | 101.6 ± 12.6 (n = 16) | n.s. | n.s. |
PS1 S367A | 99.7 ± 6.1 (n = 10) | 99.1 ± 11.4 (n = 15) | n.s. | n.s. |
PS1 S365A/S367A | 104.1 ± 6.7 (n = 19) | 102.8 ± 8.4 (n = 19) | n.s. | n.s. |
PS1 S366A/S367A | 102.7 ± 11.2 (n = 10) | 108.2 ± 9.9 (n = 19) | n.s. | n.s. |
-
*The FRET efficiency in DMSO-treated cells expressing WT PS1 is set as 100%, and relative FRET efficiency in phosphorylation-inhibited mutants of PS1 is shown. Mean ± SEM, Student’s t-test, n: cell number, †: p<0.05, n.s.: not significant.
-
‡p-value is shown for the comparison between WT PS1 and phosphorylation-inhibited mutants of PS1 in DMSO-treated conditions, or §for the comparison between DMSO-treated and A23187 (5 µM for 15 min)-treated cells expressing the same PS1 construct.
List of the human brain samples used in the study.
Case | Age | PMI | Sex | Braak | Cerad |
|---|---|---|---|---|---|
Control 1 | 60 | 15 | F | ||
Control 2 | 73 | 20 | F | ||
Control 3 | 88 | 20 | F | II | |
Control 4 | 91 | 8 | F | I | A |
Control 5 | 63 | 16 | M | ||
Control 6 | 85 | 8 | M | I | |
Control 7 | 87 | 48 | M | I | |
Control 8 | 91 | 19 | F | II | A |
Control 9 | 86 | 10 | M | ||
Control 10 | 94 | 17 | M | I | |
Control 11 | 54 | 6 | M | ||
Control 12 | 58 | 18 | F | ||
Control 13 | 88 | 16 | F | II | |
Control 14 | 60 | 14 | M | ||
Control 15 | 92 | unknown | M | II | |
Control 16 | 68 | 27 | M | I | |
Control 17 | 76 | 48 | F | I | possibly A |
Control 18 | 92 | 12 | M | II | |
Control 19 | 85 | unknown | M | II | |
Control 20 | 92 | 23 | M | II | A |
Control average (Mean ± SEM) | 79.15 ± 3.0 | 19.16 ± 2.6 | (F:M = 8:12) | ||
AD 1 | 58 | 12 | F | VI | C |
AD 2 | 73 | 18 | F | V | C |
AD 3 | 84 | 24 | F | VI | C |
AD 4 | 85 | 5 | F | VI | C |
AD 5 | 60 | 24 | M | VI | C |
AD 6 | 78 | 18 | F | VI | C |
AD 7 | 85 | 24 | M | VI | C |
AD 8 | 86 | 20 | M | VI | C |
AD 9 | 87 | 4 | F | VI | C |
AD 10 | 69 | 5 | M | VI | C |
AD 11 | 94 | 12 | F | VI | C |
AD 12 | 86 | 12 | M | VI | B |
AD 13 | 89 | 18 | F | VI | B |
AD 14 | 71 | 16 | F | VI | C |
AD 15 | 70 | 17 | M | VI | C |
AD 16 | 96 | 18 | M | VI | C |
AD 17 | 91 | 6 | M | VI | C |
AD 18 | 83 | 14 | F | VI | C |
AD 19 | 87 | 12 | F | IV | B |
AD 20 | 87 | 13 | M | VI | C |
AD 21 | 73 | 14 | M | VI | C |
AD 22 | 78 | 8 | M | VI | C |
AD 23 | 79 | 8 | M | VI | C |
AD average (Mean ± SEM) | 80.39 ± 2.1 | 14 ± 1.3 | (F:M = 11:12) |





