Functional asymmetry and electron flow in the bovine respirasome
Figures
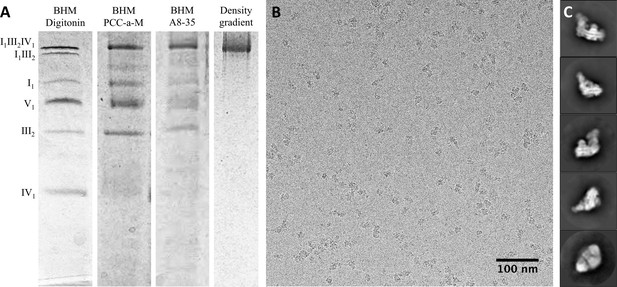
Isolation and single-particle cryo-EM of the bovine supercomplex I1III2IV1.
(A) BN-PAGE of bovine heart mitochondria (BHM) solubilized with digitonin (lane 1) or PCC-a-M (lane 2); PCC-a-M-solubilized complex after exchange to A8-35 (lane 3); density gradient fraction of supercomplex I1III2IV1 in A8-35 (lane 4). (B) Electron micrograph of bovine respirasomes in vitrified buffer, recorded with a Falcon III direct detector in integrating mode on a FEI Tecnai Polara electron microscope operating at 300 kV. (C) 2D class averages produced by reference-free 2D classification of 156,536 particles in RELION 1.4.
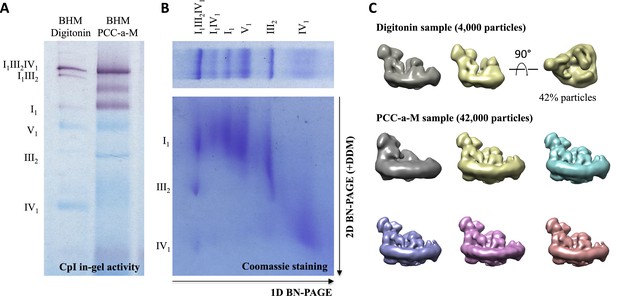
Assessment of sample quality.
(A) In-gel activity assay for NADH:dehydrogenase in BN-PAGE gel. Dark bands indicate the presence of active complex I (B) 2D BN/BN-PAGE of mitochondria solubilized with PCC-a-M confirms presence of complexes I, III and IV in the top band of the first dimension BN-PAGE lane. (C) 3D classification and reconstruction of 4000 and 42,000 negatively stained supercomplex particles after initial solubilization with digitonin or PCC-a-M. The purified digitonin-solublized sample contains a mixture of supercomplexes I1III2IV1 and I1III2, while solubilization with PCC-a-M produces pure I1III2IV1.
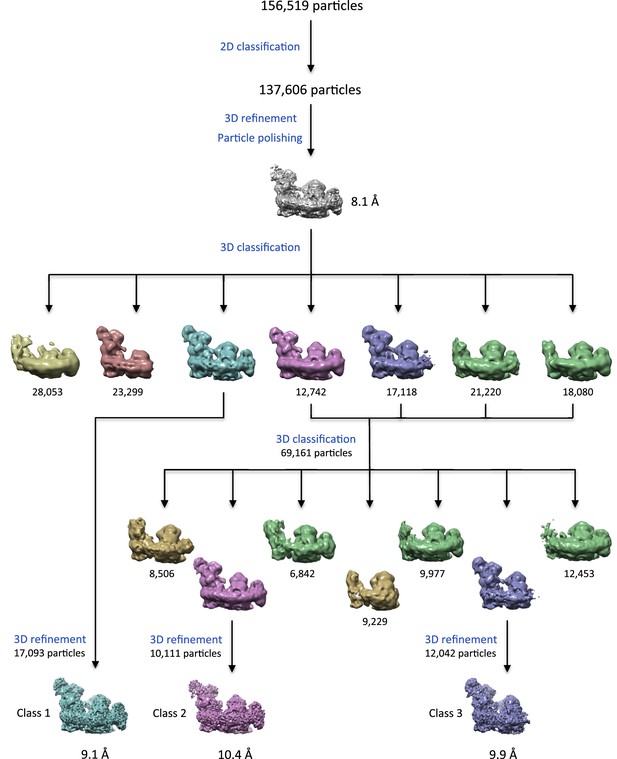
3D classification and refinement.
3D classification with RELION 1.4 reveals a range of different populations, including free complex I (salmon), I1III2IV1 lacking subunits of complex I matrix arm (green and gold) and I1III2 (violet and gold). Maps with similar features are shown in the same colors.
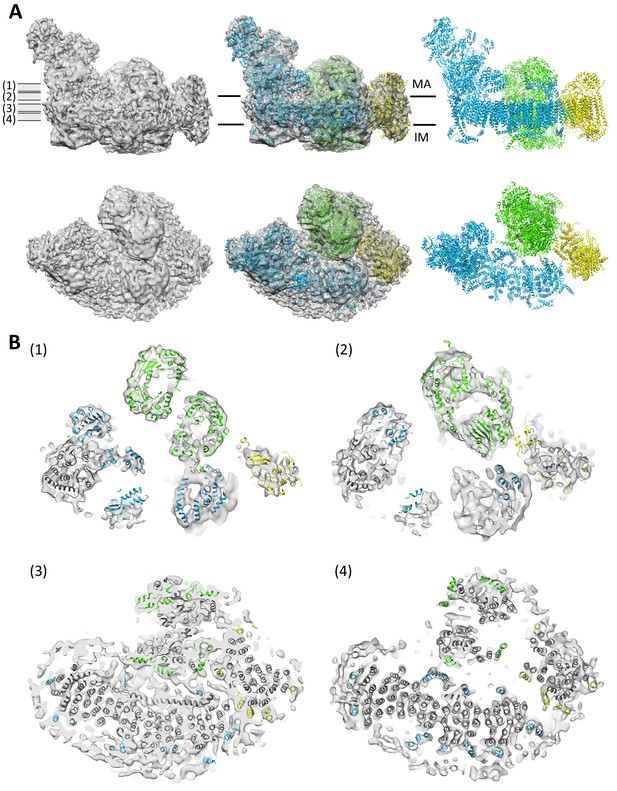
Protein-protein contacts in the respirasome.
Most contacts are mediated by supernumerary subunits of complexes I, III and IV. (A) Side views (upper row) and views from the matrix (bottom row) of class 1 cryo-EM map filtered to 8.6 Å with docked atomic models of complexes I (blue) (Vinothkumar et al., 2014), III (green) (Iwata et al., 1998) and IV (yellow) (Tsukihara et al., 1996). (B) Slices through the map at positions shown in (A) indicate contact points between the three complexes in the membrane. Core subunits are shown in grey and supernumerary subunits in color.
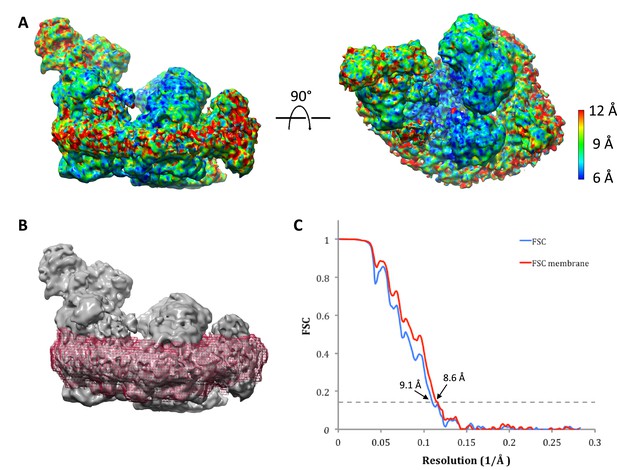
Local map resolution.
(A) Side (left) and matrix view (right) of class 1 map colored according to local resolution by RESMAP (Kucukelbir et al., 2014). The best-resolved map regions are at the interface between complexes I and III. (B) Soft mask (red) used to determine the resolution of the transmembrane domain in class 1 of supercomplex I1III2IV1. (C) Resolution estimated by gold-standard Fourier Shell Correlation (0.143 threshold). FSCs were calculated from two reconstructions obtained from half datasets of the entire map volume with a tight, soft global mask (blue) or for the masked membrane region shown in (B) (red).
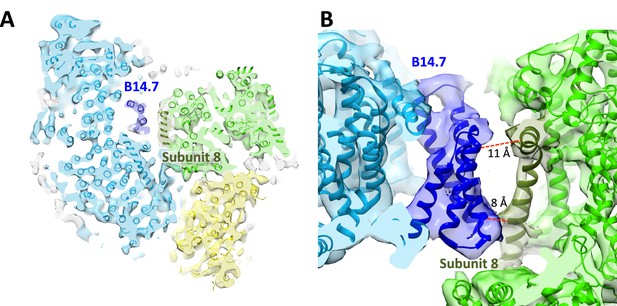
Central position of complex I supernumerary subunit B14.7.
(A) Horizontal slice through 8.6 Å map, with fitted models for complexes I (light blue), III (light green) and IV (yellow). Subunit B14.7 of complex I is dark blue and subunit 8 of complex III is dark green. (B) Detailed side view of interface shows close contacts between complex I B14.7 and complex III subunit 8 in the hydrophobic interior of the supercomplex. Distances are between α–carbons in polypeptides of adjacent complexes.
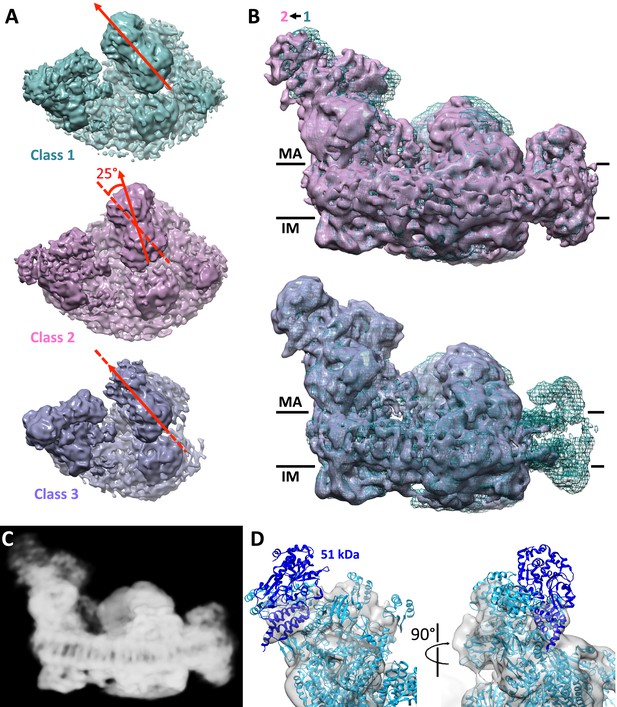
Conformational and compositional heterogeneity of the respirasome.
(A) Top view of density maps of class 1 (dark cyan) and class 2 (pink) of the respirasome and supercomplex I1III2 (violet). Classes 1 and 2 differ by a 25° rigid-body rotation of complex III in the membrane plane relative to complex I. Complex IV occupies identical positions in both maps. The arrangement of complex I and III in class 3 is similar to that in class 1. (B) Side views of classes 2 and 3 overlayed with the class 1 map (dark cyan mesh) indicate flexibility of the complex I matrix arm. In class 2 the matrix arm is displaced by 3–4° away from the center of the map. (C) Consensus map of the respirasome at 8.1 Å with weak density at the distal part of the matrix arm. (D) Complex I matrix arm in the consensus map with fitted atomic model of complex I with the 51 kDa subunit in dark blue seen from the front (left) and side (right).
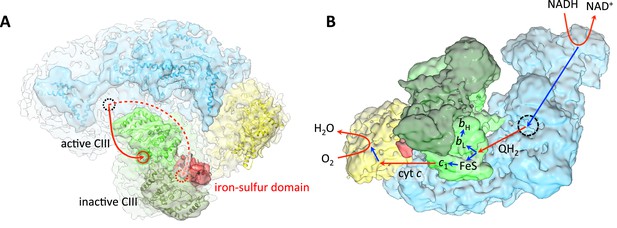
Substrate and electron flow in the respirasome.
(A) Respirasome map with fitted atomic models (blue, complex I; green, complex III; yellow, complex IV) seen from the crista lumen. Only one of the two Rieske iron-sulfur domains of complex III is resolved (red), indicating that the complex III monomer associated with it is inactive. The active complex III monomer is light green, the inactive monomer is dark green. The red circles indicate the position of the ubiquinol binding sites of complex III in the intermembrane side of the membrane. The dashed black circle indicates the position of the quinol binding site of complex I on the matrix side. (B) Oblique view from the lumenal side of the surface-rendered respirasome map, with routes of electron and substrate transfer. Red and blue straight arrows indicate substrate and electron transfer, respectively. Respiratory chain reactions are indicated by curved arrows. Electrons from NADH pass through complex I and reduce quinone to quinol in the quinol binding site (dashed black circle). Reduced quinol diffuses in the membrane, binding preferentially to the QP site of the central, active monomer of complex III. Quinol is oxidized to quinone, transferring one electron to heme bL for quinone reduction at the QN site, and another electron to the iron-sulfur cluster of the Rieske protein in the b position. The iron-sulfur domain moves from the b to the c1 position and reduces heme c1. Heme c1 reduces cytochrome c, which diffuses to complex IV where it donates electrons for reduction of O2 to H2O.





