In vivo vizualisation of mono-ADP-ribosylation by dPARP16 upon amino-acid starvation
Figures
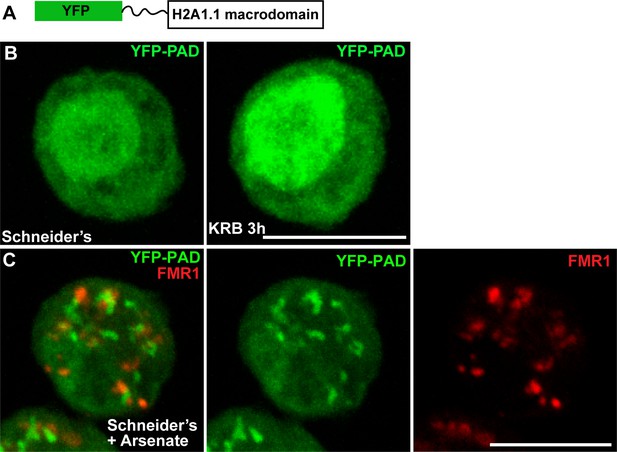
PAD in cellular stress.
(A) Schematics of YFP-PAD probe. (B) YFP-PAD in growing (Schneider’s) and amino-acid starved (KRB) cells for 3 hr. Note that with the exception of an increase of the nuclear intensity, amino-acid starvation does not lead to the formation of a cytoplasmic pattern. (C) YFP-PAD in S2 cells upon arsenate treatment. Note the formation of a robust YFP-PAD cytoplasmic pattern that co-localises with stress granules (FMR1, red). Scale bars: 10 μm
-
Figure 1—source data 1
List of the primers used in this manuscript.
- https://doi.org/10.7554/eLife.21475.003
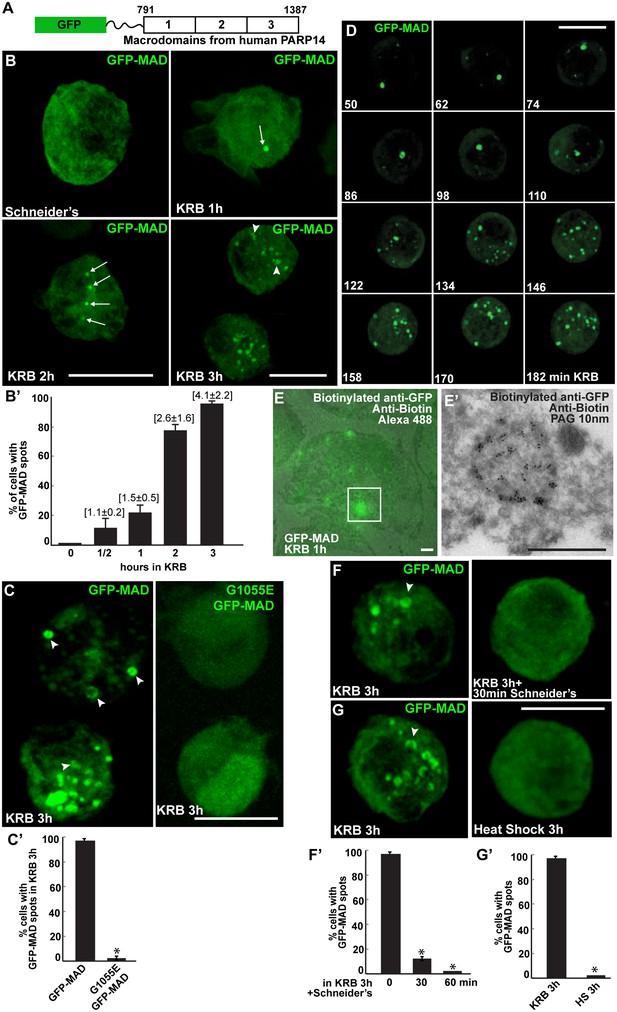
Amino-acid starvation leads to the formation of MARylation spots visualised with GFP-MAD.
(A) Schematics of the GFP-MAD probe. (B, B’) Fluorescence of GFP-MAD in growing S2 cells (Schneider’s) and upon amino-acid starvation (KRB) for increasing amount of time as indicated (B). Note the formation of GFP-MAD spots (arrows) (some in an U-shape, arrowheads in B-D). The % of cells at each time point displaying GFP-MAD spots is shown in B’. The average number of spots per cell is indicated above each bar. (C, C’) Fluorescence of GFP-MAD and G1055E GFP-MAD probe (that does not bind mono-ADP-ribose in vitro). Note that the mutant probe does not form spots in KRB (quantified in C’). (D) Stills of a time-lapse movie (Video 1) of GFP-MAD in cells incubated in KRB for 3 hr. The first frame is taken after 50 min incubation. The subsequent frames are taken every 12 min. (E, E’) Correlative Fluorescence/IEM of GFP-MAD spots in S2 cells upon amino-acid starvation (KRB, 1 hr). The IEM (E’) corresponds to the white rectangle in fluorescence that is overlapped with the corresponding electron micrograph (E). (F, F’) Fluorescence pattern of GFP-MAD and endogenous Sec16 (red) in KRB and in KRB followed by incubation with Schneider’s for 30 min. Note that GFP-MAD pattern is completely reverted (quantified in F’). (G, G’) Fluorescence pattern of GFP-MAD in KRB and upon heat shock (3 hr at 37°C). Scale bars: 10 μm (B, C, D, F, G); 500 nm (E, E’). Error bars: SEM.
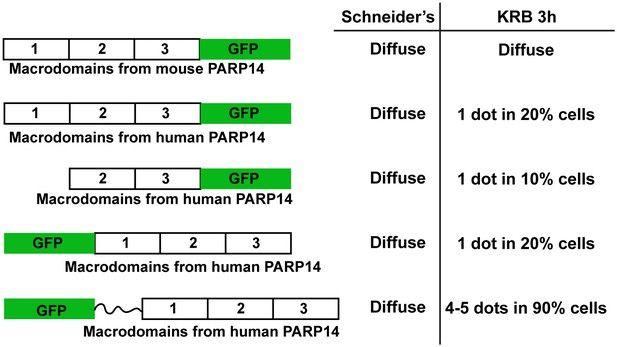
GFP-MAD design and optimisation.
Schematics representation of several version of GFP-MAD based upon macrodomains 1–3 of PARP14. Note that the macrodomains of human PARP14 are more efficient at detecting MARylation in S2 cells than those from mouse PARP14 that were originally used by (Forst et al., 2013). Furthermore, the insertion of a linker greatly improved the probe sensitivity and/or efficiency.
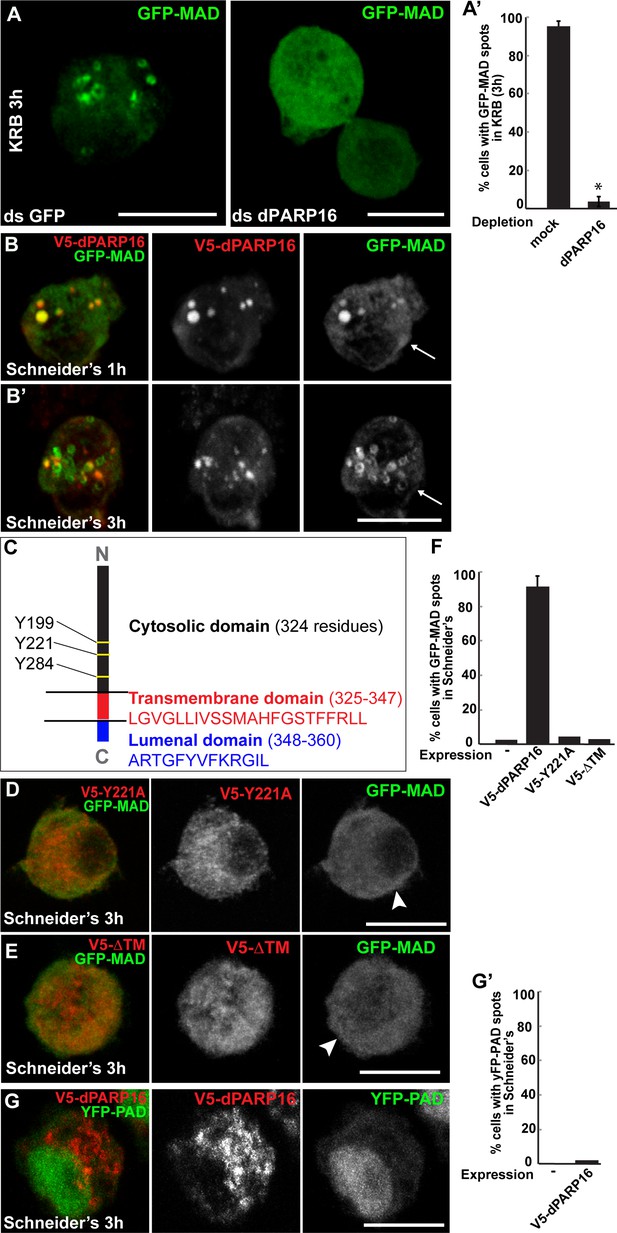
Amino-acid starvation triggered MARylation events are dPARP16 dependent.
(A, A’) Visualisation of GFP-MAD in mock and dPARP16 depleted cells upon amino-acid starvation (KRB) (A). Note that dPARP16 depleted cells do not exhibit GFP-MAD spots (quantified in A’). (B, B’) Visualisation of GFP-MAD in S2 cells expressing V5-dPARP16 in growing conditions. Note that the enzyme expression drives the formation of GFP-MAD spots in the absence of stress and that GFP-MAD spots strongly co-localise with the enzyme (quantified in F). (C) dPARP16 has 359 residues including a transmembrane domain of 22 (in red) and a luminal domain of 12 (in blue). The TM has been predicted using the TMHMM server V.2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/). The three tyrosines making up the catalytic sites are marked. (D–F) Visualisation of GFP-MAD in S2 cells expressing Y221A V5-dPARP16 catalytic mutant (E) and ΔTM V5-dPARP16 (F) in growing conditions. Note that none of the mutated forms of dPARP16 elicits GFP-MAD spot formation (arrowheads) (quantified in F). (G, G’) Visualisation of YFP-PAD in S2 cells expressing V5-dPARP16 in growing conditions. Note that the YFP-PAD localisation does not change (quantified in G’). Scale bars: 10 μm. Error bars: SEM
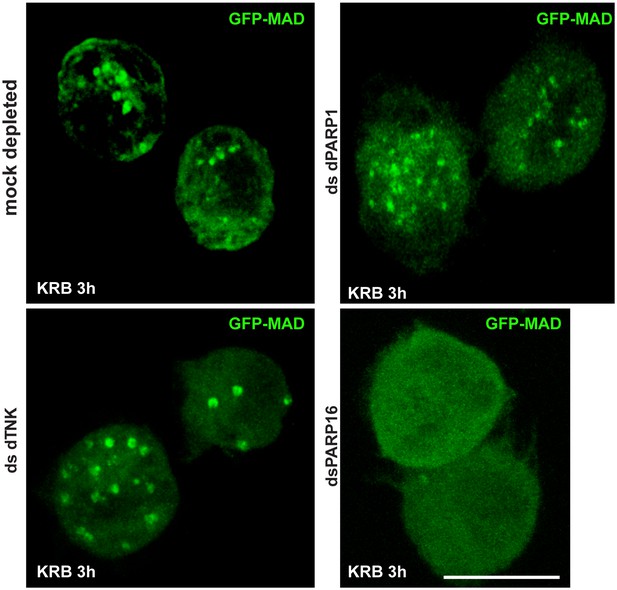
Screen for PARPs in MAD spot formation.
IF visualisation of GFP-MAD spots in amino-acid starved (KRB) mock, dPARP1, dTNK and dPARP16 depleted S2 cells. Note that, with exception of dPARP16 depletion, GFP-MAD spots formation is not affected by dPARP1 and dTNK depletions. Scale bar: 10 μm
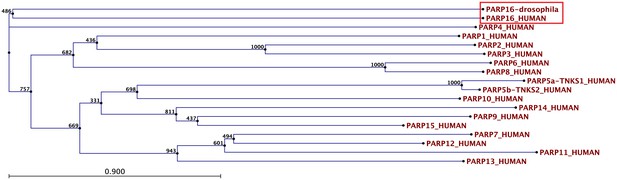
Drosophila PARP16 is the homologue of human PARP16.
Bootstrap analysis of the Drosophila PARP16 against all human PARPs. Note that dPARP16 clusters with hPARP16 (red box).

Comparison between Drosophila and human PARP16 catalytic site.
Muscle Sequence alignment of human (h) and Drosophila (d) PARP16. The yellow boxes outlines the three key amino acids that form the PARP16 catalytic site. Note that they are conserved.
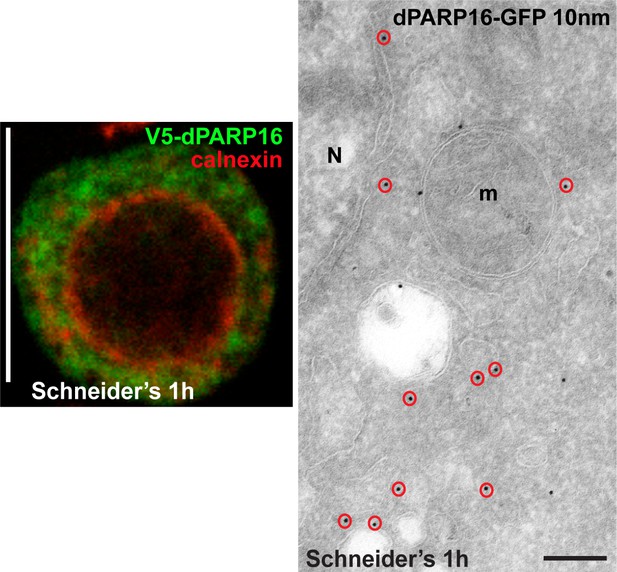
dPARP16 is anchored at the ER.
IF visualisation of V5-dPARP16 (green) with respect to the ER marker Calnexin (red) after 1 hr expression in growing S2 cells (Schneider’s). Although PARP16 only partially co-localises with calnexin, the ER localisation was confirmed by IEM (red circles). N; Nucleus. Scale bar: 10 μm (left) and 200 nm (right).
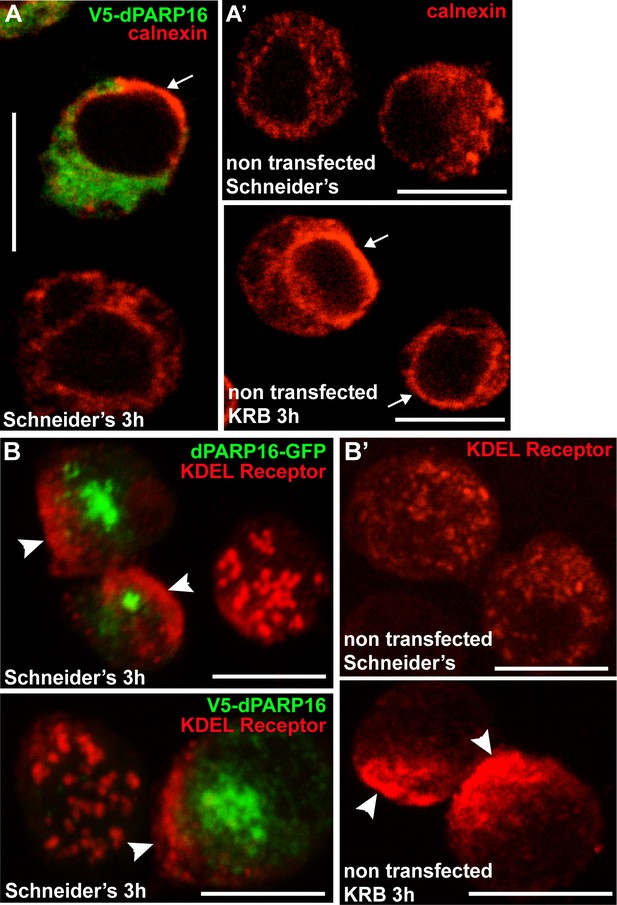
dPARP16 is anchored at the ER.
(A–B’) Overexpression of dPARP16 leads to a strong re-arrangement of the ER marked by calnexin (A) that appears to accumulate at the nuclear envelop (arrows) and KDEL receptor that appears to concentrate near the cell cortex (B, arrowheads). Interestingly, the same ER re-arrangements are observed upon amino-acid starvation for both markers (A’ and B’). Scale bars: 10 μm.
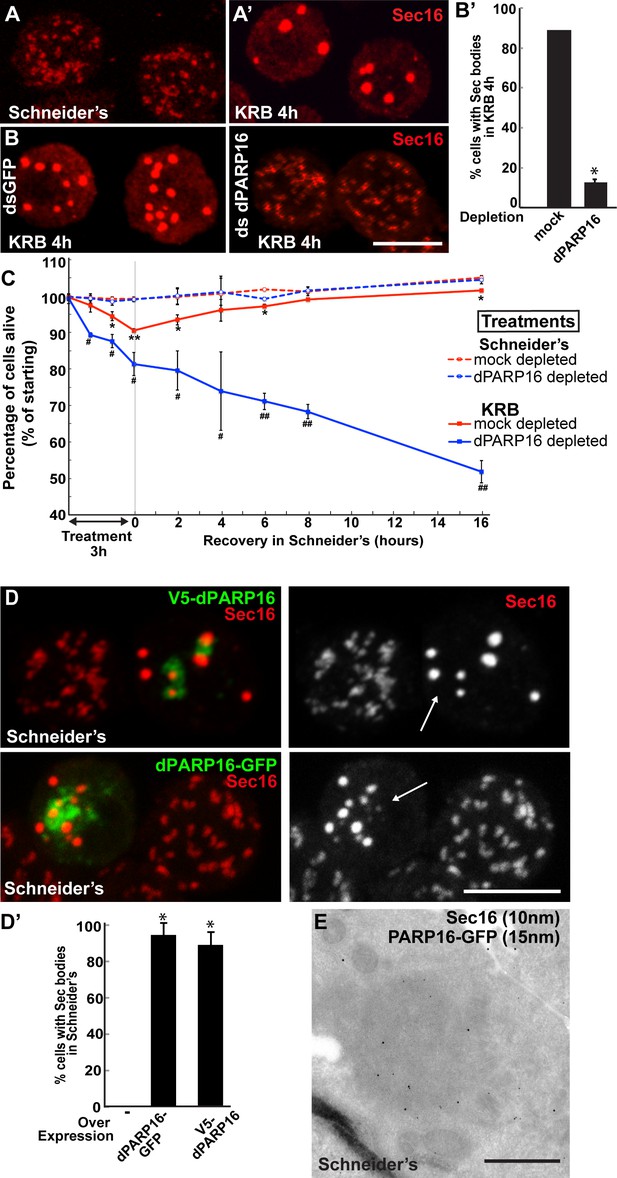
dPARP16 is required for amino-acid starvation driven Sec body formation.
(A) Immunofluorescence (IF) visualisation of endogenous Sec16 (red) at the ERES in growing Drosophila S2 cells (Schneider’s) and in Sec bodies upon amino-acid starvation (KRB). (B–B’) IF visualisation of endogenous Sec16 (red) in mock and dPARP16 depleted S2 cells upon amino-acid starvation (KRB) (B). Note that dPARP16 depletion inhibits Sec body formation (quantified in B’). (C) Graph of cell viability (expressed as percentage of alive cells) upon ‘treatments’ as indicated and recovery. The number of starting cells at t = 0, mock- (dsGFP, red lines) and dPARP16 depleted (blue lines) is set at 100%. These cells are incubated in Schneider’s (dashed lines) and KRB (solid lines) for 3 hr followed by further incubation in Schneider’s for 16 hr. Note that the dPARP16 depleted cells are more sensitive to starvation than the controls and they do not recover, whereas their viability not affected when grown in full medium. p-values were calculated for each time point corresponding to mock-depleted cells incubated in Schneider’s and in KRB. * marks p-values higher the 10−2 and **p-values higher than 10−5. p-values were also calculated for each time point corresponding to mock and dPARP16 depleted cells incubated in KRB. # marks p-values higher the 10−2 and ##, p-values higher than 10−4. (D–D’) IF visualisation of endogenous Sec16 (red) in cells) over-expressing dPARP16-GFP and V5-dPARP16 in growing cells (Schneider’s) (C). Note that it drives the robust formation of Sec bodies (arrows in C) (quantified in D’). (E) Immuno-electron microscopy (IEM) visualisation of endogenous Sec16 (10 nm gold) in dPARP16-GFP overexpressing cells (15 nm). Scale bars: 10 μm (A, B, D); 500 nm (E). Error bars: SEM (A, D’) and SD (C).
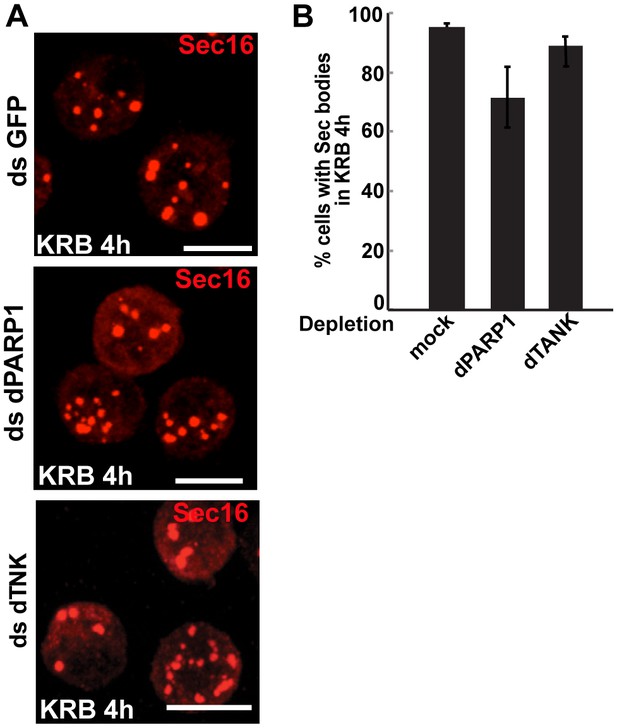
dPARP1 and dTNK depletion does not affect Sec body formation upon amino-acid starvation.
(A, B) IF visualisation of endogenous Sec16 (red) in amino-acid starved (KRB) S2 cells that are mock, dPARP1 and dTNK depleted. Note that Sec body formation is not affected as they form as efficiently in all conditions (quantified in B). Scale bars: 10 μm. Error bars: SEM.
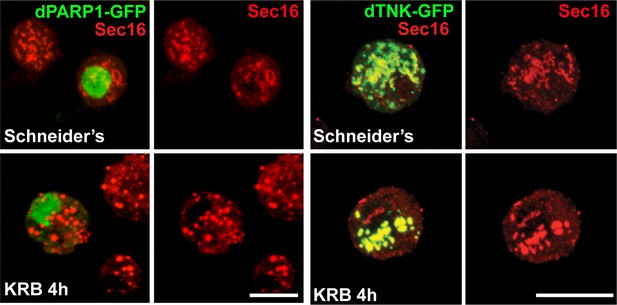
dPARP1 and dTNK overexpression does not lead to Sec body formation in growing conditions.
IF visualisation of endogenous Sec16 (red) in S2 cells expressing dPARP1 and dTNK in Schneider’s and KRB. Note that the overexpression of any of these enzymes neither leads to Sec body formation in Schneider’s nor affects their formation in KRB. Scale bars: 10 μm. Of note: dTNK, a cytoplasmic predicted MARylation enzyme, co-localises robustly with ERES in S2 cells, both under growing conditions and upon amino-acid starvation. Furthermore, its overexpression leads to the remodelling of the ERES. Yet, neither its overexpression nor its depletion has an effect on Sec body formation. This shows that the localization of MARylation enzymes at/near ERES is not sufficient to displace Sec16 and elicit a stress responsedARDT15 MARylation activity is substrate (and stress) specific.
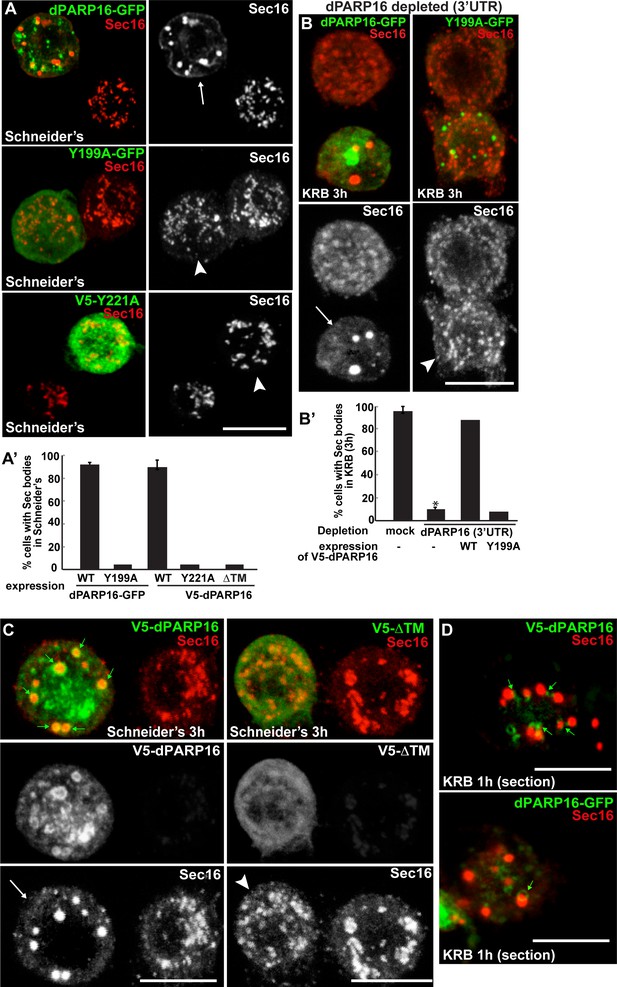
dPARP16 catalytic activity and membrane anchoring is required for Sec body formation.
(A, A’) IF visualisation of Sec body formation (Sec16, red) in growing S2 cells (Schneider’s) expressing wild type and catalytic mutant dPARP16 (Y199A and Y221A) (green) (B). Note that the expression of the catalytic mutants does not drive Sec body formation (arrowhead in B) whereas the wild type dPARP16 does (arrow in A) (quantified in A’). (B, B’) IF visualisation of Sec body formation (Sec16, red) in amino-acid starved S2 cells depleted of dPARP16 (3’UTR) and expressing wild type dPARP16 and Y199A dPARP16 catalytic mutant (B). Note that the mutant does not rescue Sec body formation (arrowhead in C) whereas the wild type dPARP16 does (arrow in B) (quantified in B’). (C) IF visualisation of Sec16 (red) upon wild type V5-dPARP16 and ΔTM V5-dPARP16 expression (green) for 3 hr in Schneider’s. Note that overexpressed wild type dPARP16 forms rings and spots (green arrows) as well as Sec bodies (white arrows) whereas the ΔTM V5 dPARP16 does not (arrowheads) (quantified in A’). (D) IF visualisation in confocal sections of V5-dPARP16 and dPARP16-GFP (green) and Sec16 (red) after 1 hr incubation in KRB. Note that the forming Sec bodies localise closely to dPARP16. Scale bars: 10 μm. Error bars: SEM.
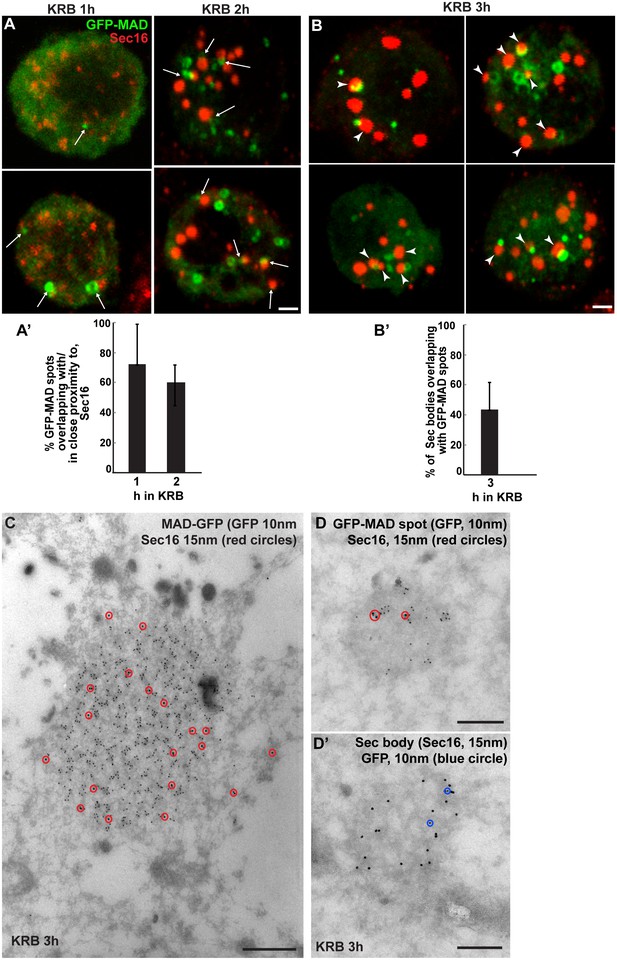
Sec bodies are formed in close proximity to GFP-MAD spots.
(A, A’) Visualisation of GFP-MAD and Sec16 (red) upon 1–2 hr incubation in KRB. The formed GFP-MAD spot are closed to, or overlap with, ERES (1 hr) and small Sec bodies (2 hr) (arrows) (quantified in A’). (B, B’) Visualisation of GFP-MAD and Sec16 (red) upon 3 hr incubation in KRB. The forming/formed Sec bodies are adjacent to MAD spots (arrowheads) (quantified in B’). (C) IEM of a GFP-MAD spot (GFP, 10 nm gold) and Sec16 (15 nm) in cells incubated in KRB for 3 hr. Note that a small pool of Sec16 is present within the GFP-MAD spots. (D–D’) IEM of GFP-MAD (10 nm gold) and Sec16 (15 nm). A small fraction of Sec16 is found in a GFP-MAD spot (D) and conversely, a small fraction of GFP-MAD is present in Sec bodies (both of the presented structures are found in the same cell). Scale bars: 1 μm (A,B) and 200 nm (C–D’). Error bars: SEM.
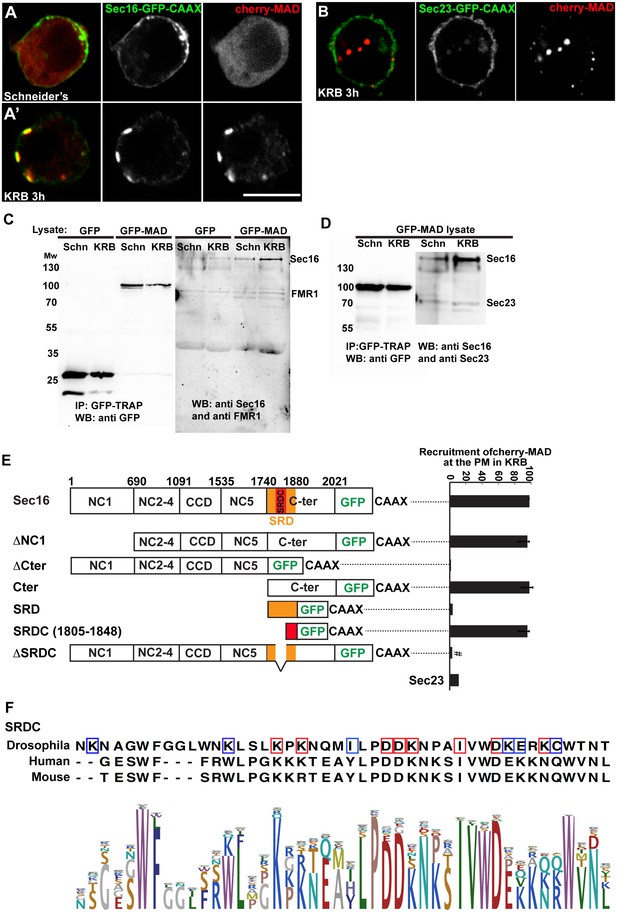
Sec16 SRCD is MARylated.
(A, A’) Co-visualisation of full length Sec16-GFP-CAAX and Cherry-MAD in growing (Schneider’s) S2 cells (A) and upon amino-acid starvation (KRB) (A’). Note that Sec16-GFP-CAAX localises to the plasma membrane where cherry-MAD is recruited upon amino-acid starvation (KRB), whereas in Schneider’s, it remains cytoplasmic. (B) Co-visualisation of full length Sec23-GFP-CAAX (B) and cherry-MAD upon amino-acid starvation (KRB). Note that cherry-MAD is not recruited to the plasma membrane and forms spots in the cytoplasm. (C) WB (using anti GFP, anti Sec16, anti FMR1) of GFP-MAD immuno-precipitation (IP) using GFP-TRAP from stable S2 cell lines expressing GFP-MAD and GFP, either in growing conditions (Schneider’s) or upon amino-acid starvation (KRB 3 hr). (D) WB (using anti GFP, anti Sec16, anti Sec23) of GFP-MAD IP from stable S2 cells expressing GFP-MAD, either in growing conditions (Schneider’s) or upon amino-acid starvation (KRB 3 hr). Note that Sec23 pull-down by GFP-MAD is very weak when compared to Sec16. (E) Map of all Sec16-GFP-CAAX truncations used and the quantitation of cherry-MAD recruitment to the plasma membrane. Note that Sec16-ΔSRDC-GFP-CAAX transfection was performed in Sec16 depleted cells (marked by #) to avoid oligomerisation with endogenous Sec16. (F) Muscle sequence alignment of Drosophila, human and mouse Sec16 SRDC (1805–1848) and presentation of logo sequence outlining the degree of sequence conservation among all eukaryotes (defined using CLC Main Workbench 6.7.1 using the full length Sec16 sequence against all eukaryote sequences from the non-redundant protein database using standard settings). The red squares indicate the conserved residues that are potentially MARylated and the blue squares the non-conserved ones.
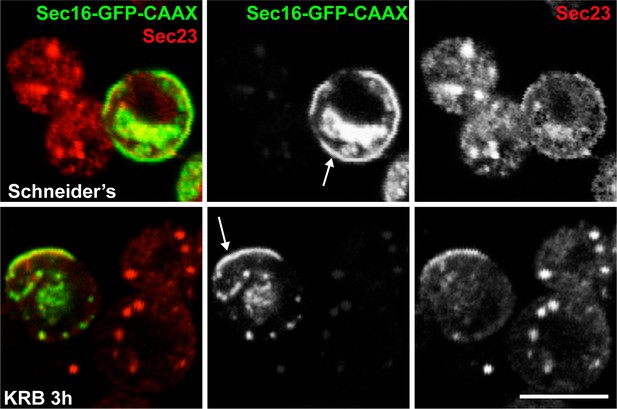
Sec16-CAAX recruits Sec23 to the PM upon AA starvation.
Co-visualisation of full length Sec16 (Sec16-GFP-CAAX) and Sec23 in growing conditions (Schneider’s) and upon amino-acid starvation (KRB). Note that Sec16-GFP-CAAX localises at the plasma membrane in Schneider’s and KRB but that Sec23 is only recruited upon KRB incubation, whereas in Schneider’s, it localises at ERES. Note also that Sec bodies do not form in the CAAX transfected cells. Scale bars: 10 μm.
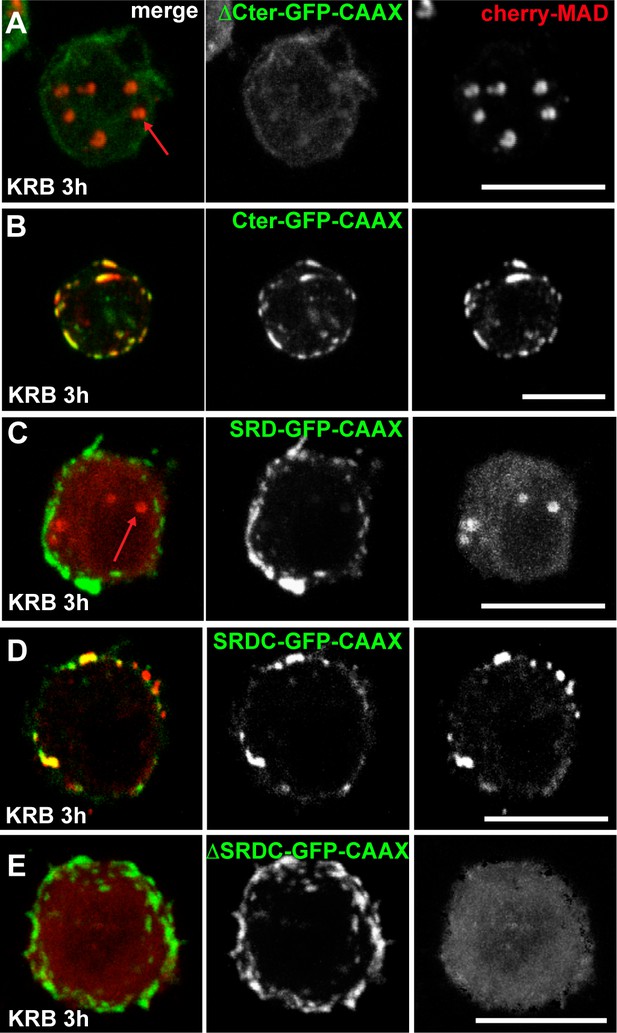
Sec16 SRDC is MARylated upon amino-acid starvation.
(A–C) Co-visualisation of Sec16 ΔCter-GFP-CAAX (A), Sec16 Cter-GFP-CAAX (B), SRD-GFP-CAAX (C) co-transfected with cherry-MAD in wild type cells in KRB. (D–E) Co-visualisation of SRDC-GFP-CAAX (D) and Sec16-ΔSRDC-GFP-CAAX (E) co-transfected with cherry-MAD in KRB (quantified in Figure 7E’). Note that although SRD does not recruit cherry-MAD, SRDC does. Scale bars: 10 μm.
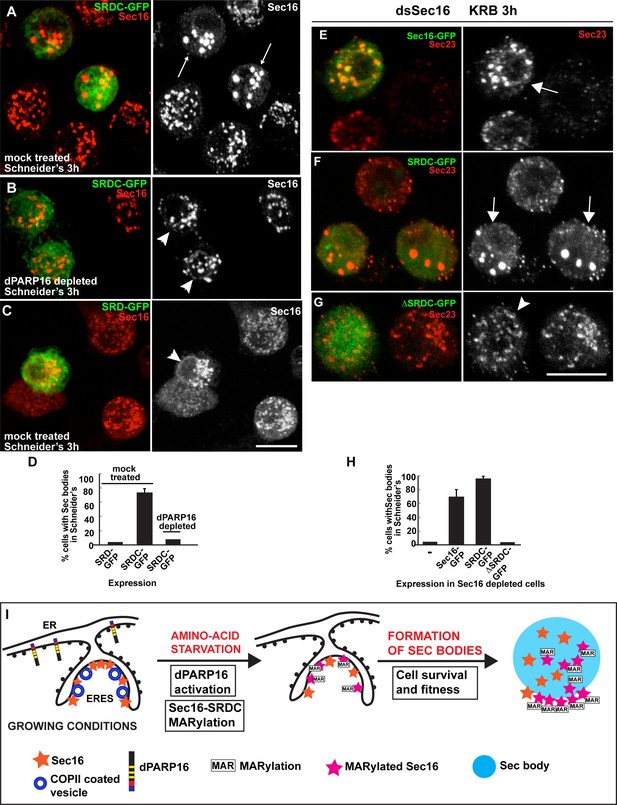
Sec16 SRDC MARylation is critical for Sec body formation.
(A–D) Visualisation of endogenous Sec16 (red) in mock treated (A,C) and dPARP16 depleted S2 cells (B) in Schneider’s transfected with the Sec domains SRDC-GFP (A,C) and SRD-GFP (B). Note that expression of SRDC leads to the formation of Sec bodies, similar to dPARP16 overexpression and that dPARP16 depletion prevents this formation (quantified in D). ( E–H) Visualisation of endogenous Sec23 (to mark Sec bodies) in starved Sec16 depleted cells (ds Sec16) transfected with Sec16-GFP (E), SRDC-GFP (F) and ΔSRDC-GFP (G). Note that Sec16 depletion prevent cells to form Sec bodies. Both, full length Sec16 and SRDC transfection rescue Sec body formation, but not Sec16 lacking SRDC. Arrows point to cells where Sec bodies have formed and arrowheads where they haven’t. (I) Model of Sec body formation upon amino-acid starvation of Drosophila S2 cells. Under growing conditions, Sec16 is at ERES where COPII coated vesicles bud. dPARP16 is at the ER. Upon amino-acid starvation, dPARP16 is activated and MARylates SRDC, a 44 amino-acid stretch in the C-terminus of Sec16. Sec16 MARylation (and possibly MARylation of other components) triggers the formation of Sec bodies by dynamic incorporation of MARylated substrates within the structure and promotes cell survival and fitness. Scale bars: 10 μm. Error bars: SEM.
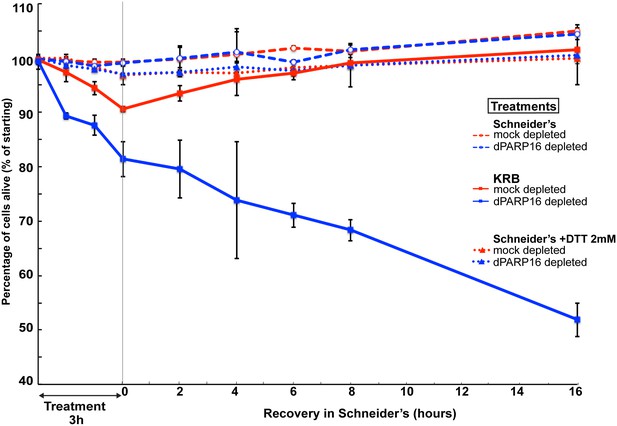
Comparison of cell viability upon amino-acid starvation and ER stress.
Graph of cell viability (expressed as percentage of alive cells) upon and recovery. The number of starting cells at t = 0, mock- (dsGFP, red lines) and dPARP16 depleted (blue lines) is set at 100%. These cells are incubated in Schneider’s (dashed lines) KRB (solid lines) and Schneider’s supplemented by 2 mM DTT (ER stress) (dotted lines) for 3 hr followed by further incubation in Schneider’s for 16 hr. For each time point, p-values were calculated between cells incubated in Schneider’s alone and Schneider’s +DTT as well as between mock and dPARP16 depleted cells treated with DTT. There were no significant difference between these samples.
Videos
GFP-MAD time-lapse movie of one cells incubated in KRB (t = 0) for 3 hr.
One frame was taken every 10 min and the movies are displayed at 12 frame/s (related to Figure 2D).





