Structural insights into the mechanism of the DEAH-box RNA helicase Prp43
Figures
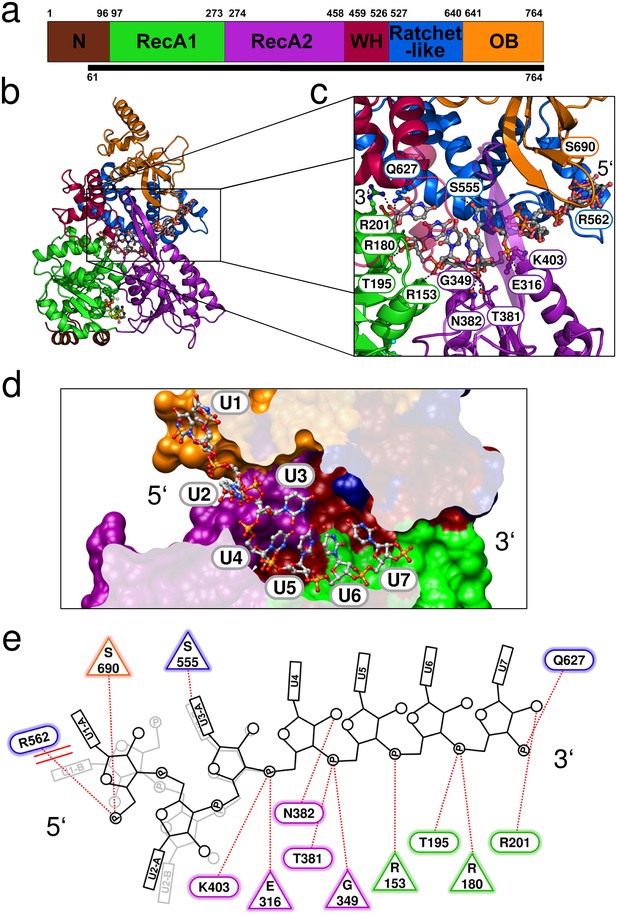
Crystal structure of Prp43 in complex with U7-RNA and the ATP-analog ADP•BeF3-.
(a) Domain overview of ctPrp43. The bottom bar indicates the N-terminally truncated construct (ΔN, 61–764) used for crystallization. (b) Overall structure of ctPrp43ΔN•U7•ADP•BeF3-. Domains are colored according to a and the bound U7-RNA is shown in gray with two alternative conformations for nucleotides U1-U3. The ADP•BeF3- is bound in the cleft between the RecA domains. (c) Close-up of the bound U7-RNA. Residues involved in interactions are labeled according to the wild-type ctPrp43 sequence. The 5’ and 3’ end of the RNA is indicated. (d) Cross-section of the Prp43 RNA-binding tunnel. Prp43 is shown in surface representation and the RNA in ball-and-sticks mode. (e) Schematic figure of the Prp43-RNA interactions. Residues which interact with the RNA via their main chain are shown as triangles and residues which exhibit side chain interactions are presented as ellipses. The coloring of the residues corresponds to a. The alternative conformation of the first three nucleotides is shown light gray. Stacking interactions are highlighted by double lines and polar interactions by dotted lines.
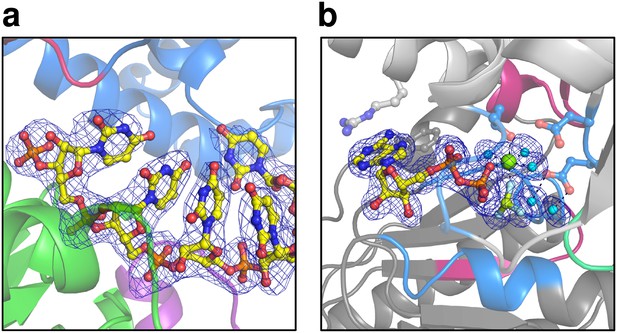
Omit maps of a fraction of the U7-RNA from the ctPrp43ΔN•U7•ADP•BeF3- complex structure and of the active site of ctPrp43ΔN•ADP•BeF3-(HR) (Figure 2a).
(a) |Fo-Fc| omit map of nucleotides U3-U7 at 3σ level. The complete RNA molecule was omitted for map calculation. Coloring according to Figure 1a. (b) The ADP•BeF3-, the Mg2+ ion and five water molecules at the active site were omitted during map calculation. The |Fo-Fc| map is shown at 3σ. Coloring according to Figure 6a.
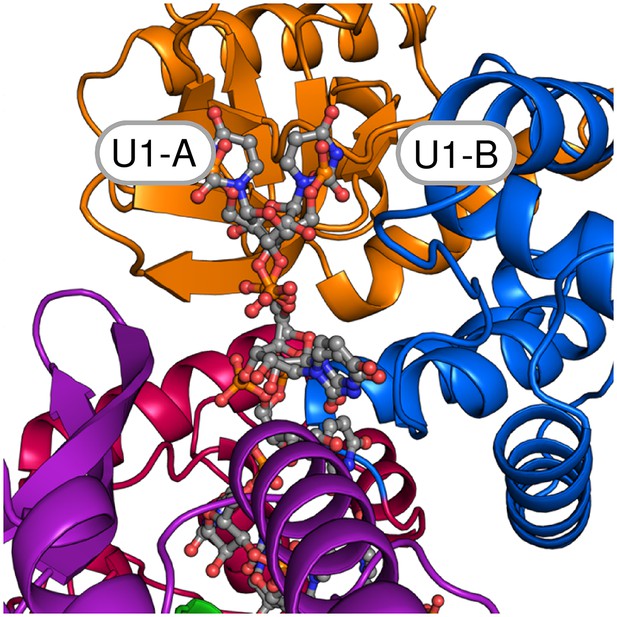
The two alternative conformations of the U7-RNA in the ctPrp43ΔN•U7•ADP•BeF3- complex structure.
Coloring according to Figure 1a. Nucleotides U1-U3 are present in two alternative conformations (A and B) which exhibit after crystallographic refinement an occupancy of 54% and 46%, respectively. The first nucleotide of each conformation is labeled.
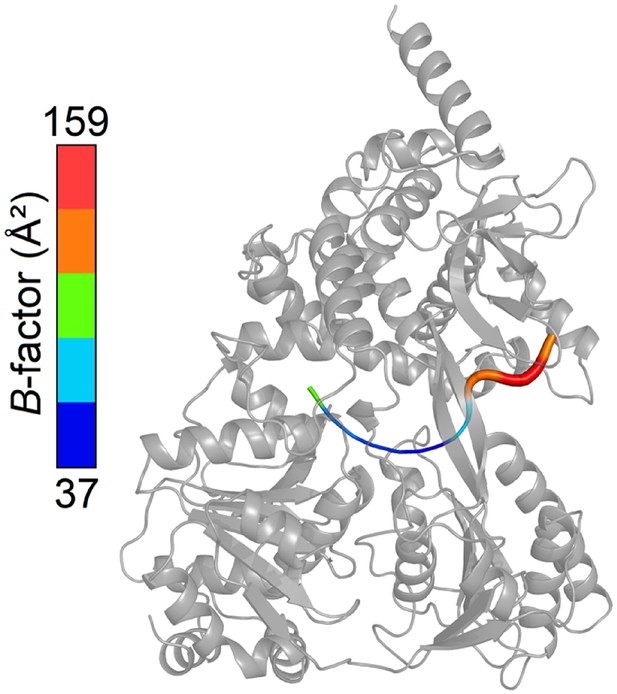
Overview of the B-factors of the U7-RNA in the ctPrp43ΔN•U7•ADP•BeF3- complex structure.
B-factors of the bound RNA range from 37 to 159 Ų. The B-factors of U1 to U3 are clearly elevated compared to U4 to U7.
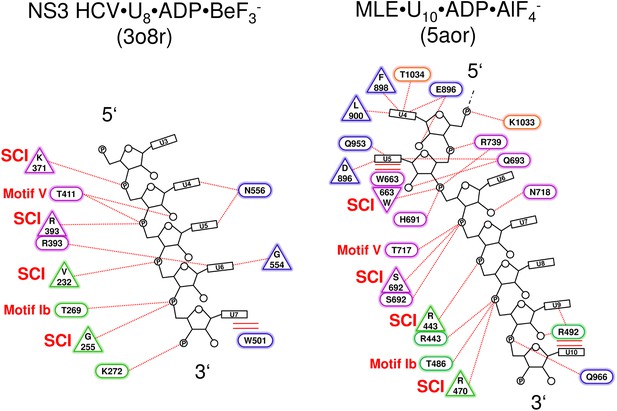
Schematic representation of the NS3 HCV- and MLE-RNA interaction networks.
(a) Interactions between NS3 HCV•ADP•BeF3- (PDBid: 3o8r) and the bound U8-RNA are shown. Polar interactions are presented as dotted lines and stacking interactions as double lines. Residues which interact via their main chain are depicted as triangles and residues which interact with their side chain are shown as ellipses. RecA1 domain residues are labeled in green, RecA2 residues in purple and Domain 3 residues in blue. The 5’ and 3’ end of the RNA is labeled according to convention. Structurally conserved interactions (SCI) between main chain amides and the RNA which are also present in Prp43 are indicated as well as the conserved threonine side chain interactions from motif Ib and V. (b) Depiction of the MLE•ADP•AlF4- (PDBid: 5aor) and U10-RNA interaction network. Residues from the RecA1 are shown in green, RecA2 residues in purple, ratchet-like domain residues in blue and residues which are located in the OB-fold are presented in orange. Abbreviations were used as introduced in a. The first three nucleotides (U1–U3) present in the crystal structure of MLE were omitted in this figure, since these interactions are neither conserved in Prp43 nor in NS3 HCV.
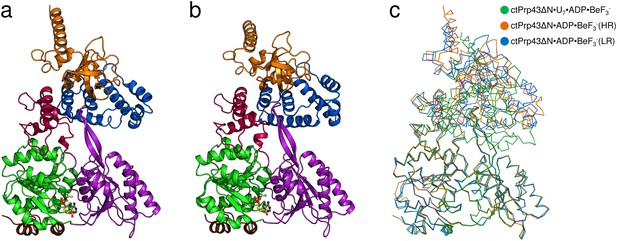
Structures of Prp43 with the bound ATP-analog ADP•BeF3- in different crystal forms.
(a) Overall structure of ctPrp43ΔN•ADP•BeF3- at high and (b) low resolution. ctPrp43ΔN•ADP•BeF3-(HR) crystalized in the orthorhombic space group P212121 and ctPrp43ΔN•ADP•BeF3-(LR) in the hexagonal space group P65. Domains are colored according to Figure 1a. ADP•BeF3- is shown at the binding cleft between the two RecA-like domains in ball-and-stick mode. (c) Superposition of ctPrp43ΔN•ADP•BeF3-(HR) (orange) and ctPrp43ΔN•ADP•BeF3-(LR) (blue) with ctPrp43ΔN•U7•ADP•BeF3- (green). Structures were superimposed via their helicase core (RecA1 and RecA2 domains) and are shown in ribbon representation.
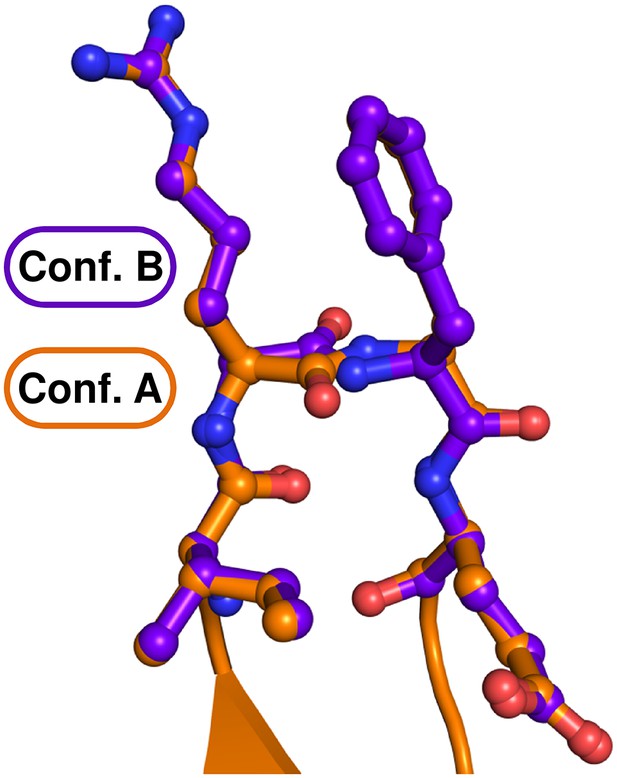
The main chain of the Hook-Turn (RF motif) is present in two alternative conformations.
The A conformation (47%) is shown in orange and the B conformation (53%) in purple blue. In the A conformation a type I β -turn is formed and in the B conformation a type II β-turn.
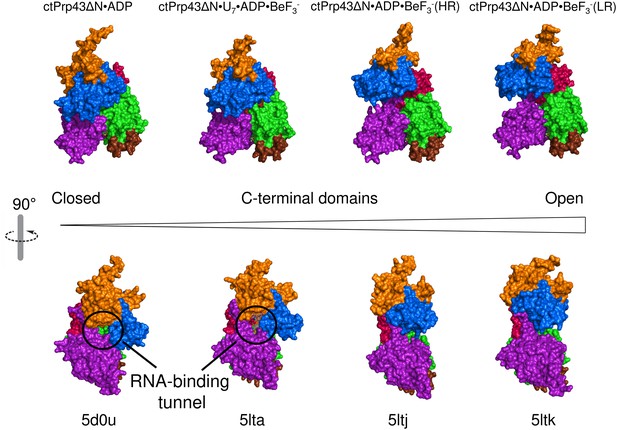
Conformational changes between the ctPrp43ΔN•ADP complex structure (5d0u) (Tauchert et al., 2016), ctPrp43ΔN•U7•ADP•BeF3- (5lta), ctPrp43ΔN•ADP•BeF3-(HR) (5ltj) and ctPrp43ΔN•ADP•BeF3-(LR) (5ltk).
Structures were superposed via their RecA1 domains. In the top panel the back view is presented (rotated by 180° with respect to Figure 1b, Figure 2a and Figure 2b) and in the bottom panel the side view (90° rotation around a vertical axis). Structures are ordered according to the degree of C-terminal displacement and opening of the RNA-binding tunnel. The location of the RNA-binding tunnel which is present in ctPrp43ΔN•ADP and ctPrp43ΔN•U7•ADP•BeF3- is indicated.
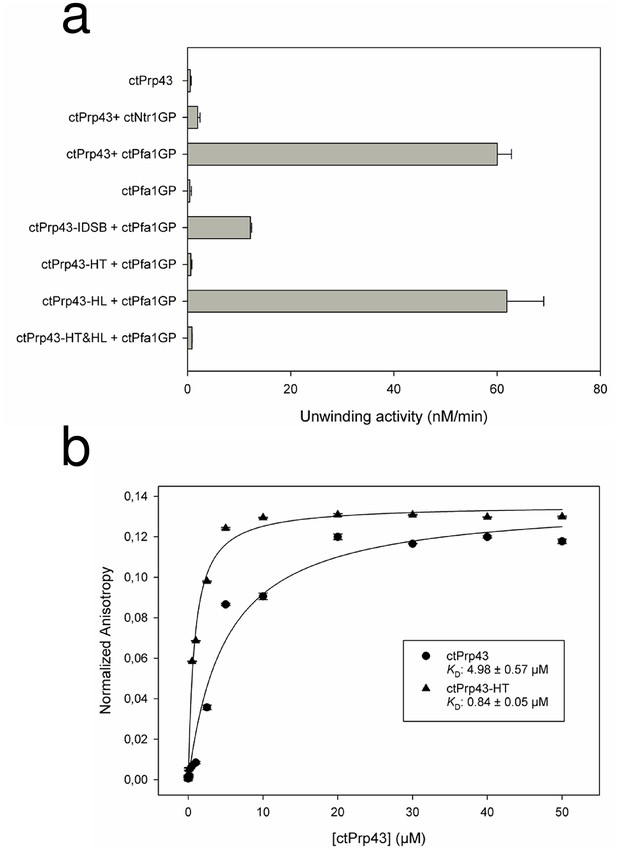
Helicase activity and RNA-binding assays of ctPrp43 and mutants.
(a) The maximal unwinding velocity (nM/min) for a dsRNA with a 3’ overhang is shown. (b) RNA binding of 5’−6FAM-U16-RNA by ctPrp43 and ctPrp43-HT was determined via fluorescence anisotropy measurements. Error bars indicate the standard deviation from three independent measurements for a and b.
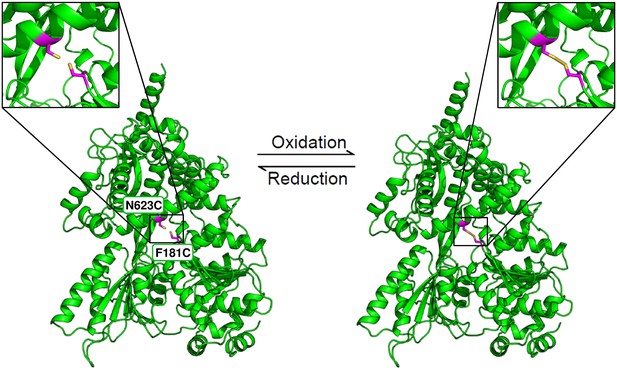
Position of the two introduced cysteine residues in the ctPrp43-IDSB mutant.
The Phe 181 in the RecA1 domain and the Asn 623 in the ratchet-like domain were substituted by cysteines. Owing to their close spatial proximity, a disulfide bridge can be formed which traps Prp43 in this closed conformation. The formation of this disulfide bridge is reversible, for instance, by adding a reducing agent such as DTT. The closed state of the ctPrp43-IDSB mutant was modeled from the ctPrp43ΔN•U7•ADP•BeF3- complex.
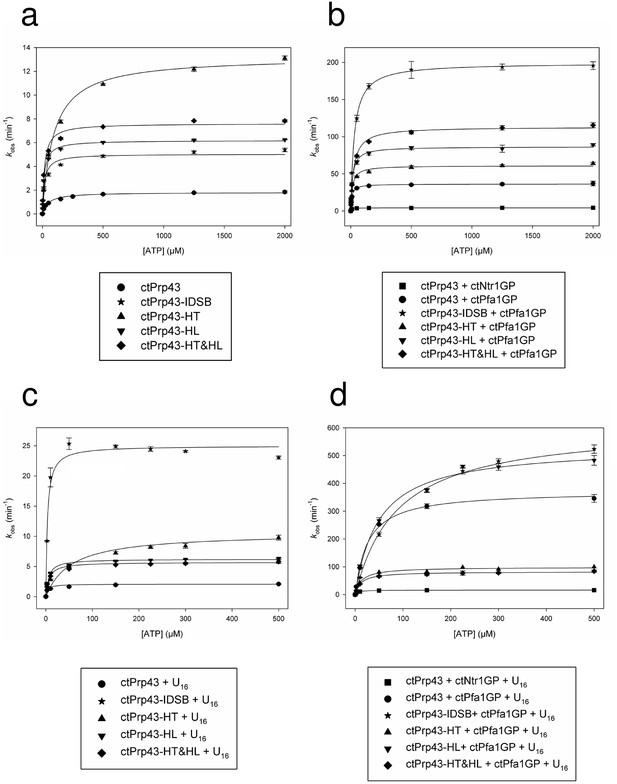
ATPase activity of Prp43 and mutants (a) without further stimulation, (b) in the presence of a G-patch, (c) in the presence of a U16-RNA and (d) in the presence of a G-patch and a U16-RNA.
kcat and KM values are shown in Table 3. Error bars, s.d. (n = 3 independent measurements). For detailed information refer to the Materials and methods section.
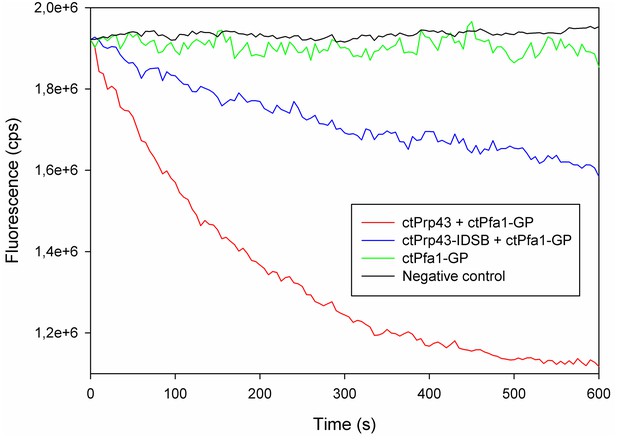
Raw data of exemplary helicase activity measurements.
The typical curve progression of individual measurements for this fluorescence-based helicase assay is shown for ctPrp43+ctPfa1-GP (red), ctPrp43-IDSB+ctPfa1-GP (blue), ctPfa1-GP (green) and the negative control (black) in which no proteins were present.
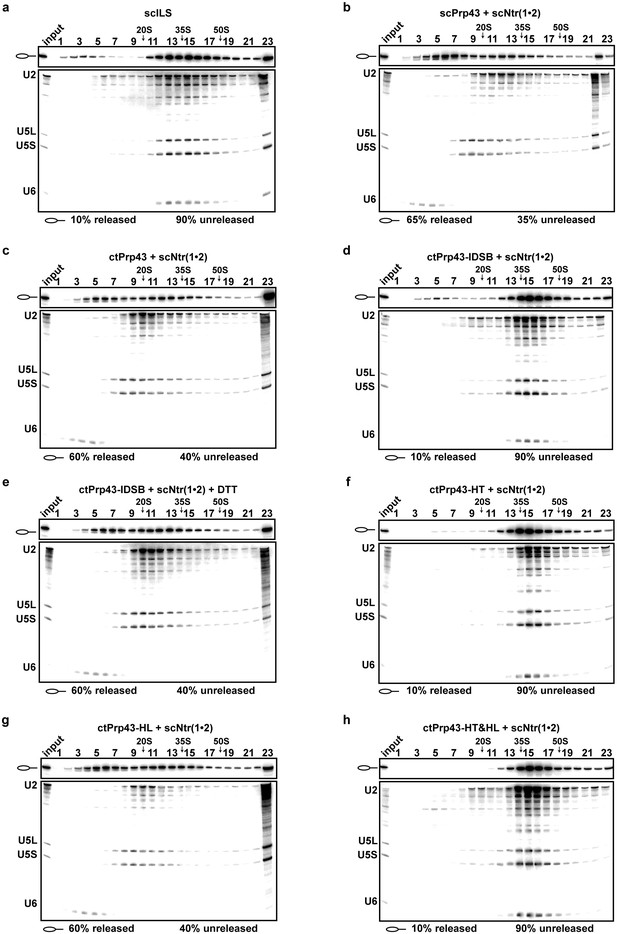
Intron-lariat spliceosome (ILS) disassembly assays.
10–30% glycerol gradient sedimentation of purified yeast ILS (scILS) incubated in solution with ATP plus (a) no recombinant protein, (b) scPrp43 and cofactors scNtr(1•2), (c) ctPrp43 and scNtr(1•2), (d) ctPrp43-IDSB and scNtr(1•2), (e) ctPrp43-IDSB, scNtr(1•2) and 0.5 mM DTT, (f) ctPrp43-HT and scNtr(1•2), (g) ctPrp43-HL and scNtr(1•2), (h) ctPrp43-HT&HL and scNtr(1•2). U2, U5 and U6 snRNAs were visualized by Northern blotting followed by autoradiography. RNA identities are indicated on the left. Quantifications were performed with ImageQuant software (Molecular Dynamics). Numbers represent the percentage of intron-lariat RNA released in the top fractions (sum of fractions 1–11) or associated with the ILS (unreleased, sum of fractions 12–23) relative to the intron-lariat RNA distributed in all 23 fractions, the sum of which was set to 100%.
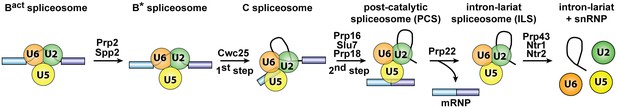
Isolation of intron-lariat spliceosomes (ILSs).
Activated spliceosomes (BactΔPrp2) assembled on Actin7 wild-type pre-mRNA in heat-inactivated splicing extracts from a prp2-1 yeast strain expressing a temperature-sensitive Prp2 mutant, were first purified. Purified BactΔPrp2 complexes were then incubated with recombinant Prp2 and Spp2, generating the B* spliceosome, and then Cwc25 was added to promote catalysis of step 1 of splicing and the formation of complex C. For catalysis of step 2, which generates post-catalytic spliceosomes (PCS), recombinant Prp16, Slu7 and Prp18 were added. Finally, for the purification of the ILS, the spliced mRNA was dissociated from the ILS by incubation of the PCS with Prp22 and ATP. Addition of ATP and recombinant Prp43, Ntr1 and Ntr2, leads to disassembly of the ILS into the intron-lariat, 20S U2 snRNP, 18S U5 snRNP and free U6 snRNA.
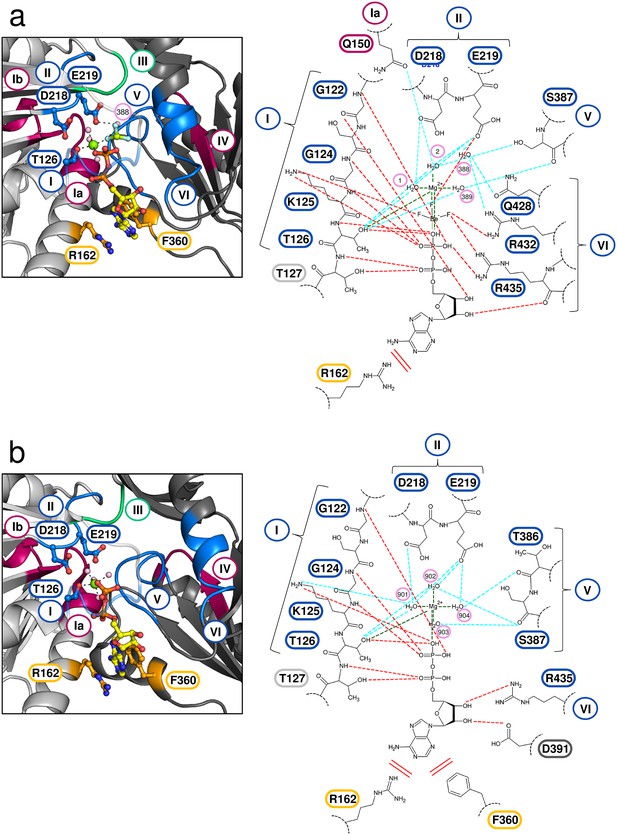
Active site of Prp43 in the ATP- and ADP-bound state.
(a, Left panel) Active site of Prp43 with the bound ATP-analog ADP•BeF3- as present in ctPrp43ΔN•ADP•BeF3-(HR) (PDBid: 5ltj). The RecA1 domain is colored in light gray, the RecA2 domain in dark gray, carbon atoms of the ADP in yellow, oxygen in red, nitrogen in blue, phosphorus in orange, beryllium in chartreuse, fluoride in light blue, magnesium in light green and water molecules in pale pink. Nucleotide-interacting motifs are shown in blue, nucleic acid-binding motifs in ruby and motif III, which couples ATP hydrolysis to RNA unwinding, in green. Residues, which are involved in base-stacking with the adenine moiety, are presented in light orange. Residues from motif I, which are involved in Mg2+ coordination (Thr 126), and from motif II, which coordinate water molecules at the active site (Asp 218 and Glu 219), are presented as sticks. The water molecule 388 is perfectly positioned for the nucleophilic attack on the γ-phosphate and thus its hydrolysis. The conserved SF2 helicase motifs I-VI are numbered according to convention. (a, Right panel) Schematic representation of the active site in ctPrp43ΔN•ADP•BeF3-(HR). Motifs and residues are labeled as introduced in the left panel. Water molecules are numbered according to the PDB entry. Stacking interactions are shown by double lines, polar interactions between ADP•BeF3- and Prp43 via red dotted lines, interactions between Prp43 and water molecules via light blue lines and the coordination of the central Mg2+ by dark green lines. (b) Active site of Prp43 with bound ADP (PDBid: 5d0u) (Tauchert et al., 2016). (Left and right panel) Labeling and numbering according to a. Numerous rearrangements and conformational changes are noticeable between the ATP- and ADP-bound state.
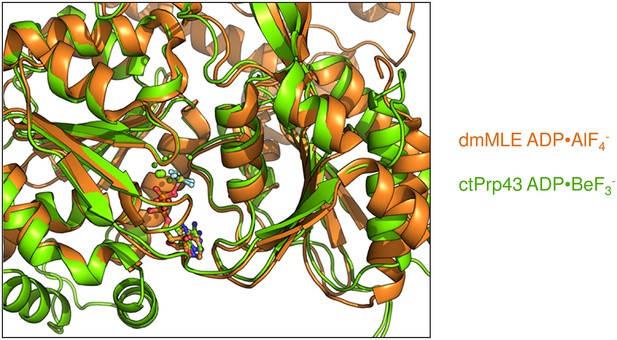
Superposition of the RecA-like domains of the ctPrp43ΔN•ADP•BeF3- (HR) structure with chain A of the MLE•U10•ADP•AlF4- complex (PDB 5aor) (Prabu et al., 2015).
The conformation of the helicase core is highly similar, also for all conserved helicase SF2 motifs, indicated by an r.m.s.d. value of 0.70 Å for 235 Cα for the superposition.
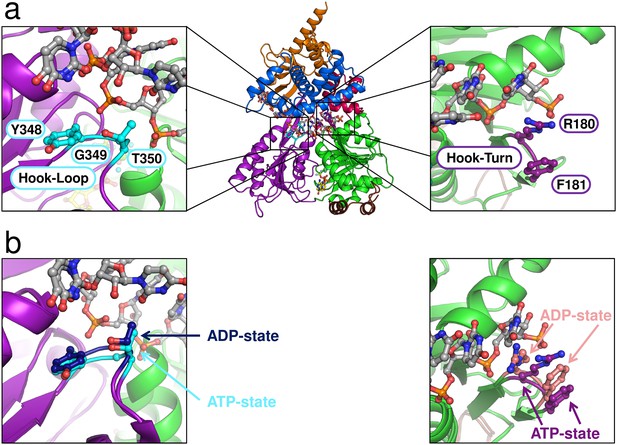
Position of the Hook-Turn and Hook-Loop in ctPrp43.
(a) The localization of the Hook-Turn in the RecA1 domain and of the Hook-Loop in the RecA2 domain in the ctPrp43ΔN•U7•ADP•BeF3- complex structure is shown. Domains are colored according to Figure 1a. (b) Superpositions of the RecA1 and RecA2 domains of the ctPrp43ΔN•U7•ADP•BeF3- and the ctPrp43ΔN•ADP (PDB 5d0u) complexes for the Hook-Turn and Hook-Loop, respectively. The superpositions indicate that the Hook-Loop remains in a highly similar conformation after ATP hydrolysis in contrast to the Hook-Turn which is shifted towards the RNA in the ADP-bound state.
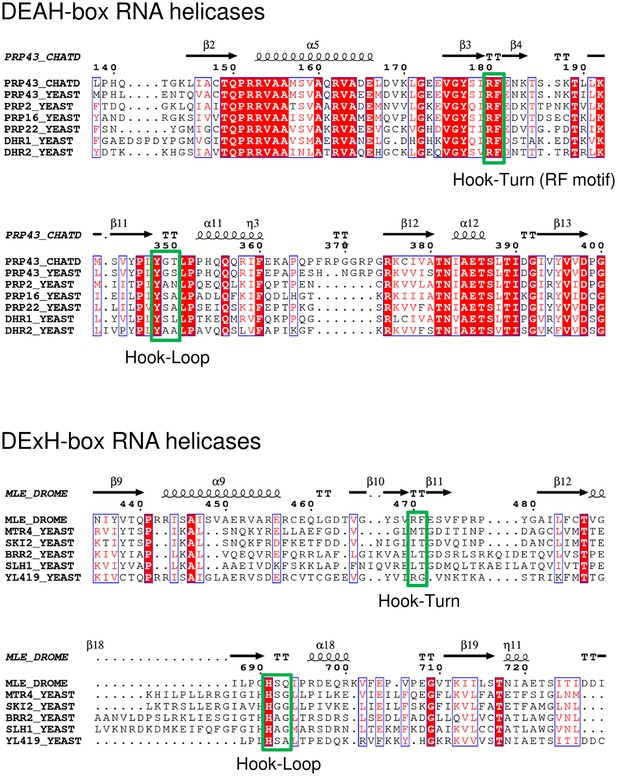
Partial sequence alignment of Prp43 from C. thermophilum to all DEAH-box RNA helicases from S. cerevisiae and of MLE from D. melanogaster to all yeast DExH-box RNA helicases.
The amino-acid sequence of ctPrp43 (G0RY84) was aligned to scPrp43 (P53131), scPrp2 (P20095), scPrp16 (P15938), scPrp22 (P24384), scDhr1 (Q04217) and scDhr2 (P36009). The sequence of dmMLE (P24785) was aligned to scMtr4 (P47047), scSki2 (P35207), scBrr2 (P32639), scSlh1 (P53327) and scYL419 (Q06698). The values in parentheses indicate the Uniprot accession numbers (The UniProt Consortium, 2015). The alignment was visualized with ESPript 3.0 (Robert and Gouet, 2014). The Hook-Turn and Hook-Loop motifs are boxed in green.
Videos
Prp43 adopts an open conformation after ATP binding and switches into the closed conformation after binding to RNA.
This morphing movie between the ADP-bound state of Prp43 (PDBid: 5d0u), the two ADP•BeF3- bound states (PDBids: 5ltk and 5ltj) and the ctPrp43ΔN•U7•ADP•BeF3- complex structure (PDBid: 5lta) illustrates the RNA-binding mode of Prp43.
Local conformational rearrangements at the active site induce global conformational changes in Prp43 which are coupled with the unwinding activity.
The morphing between the ctPrp43ΔN•U7•ADP•BeF3- (PDBid: 5lta) complex structure and ctPrp43ΔN•ADP (PDBid: 5d0u) reveals conformational rearrangements in the course of ATP hydrolysis, mainly of the Hook-Turn, which is in close spatial proximity to the RNA.
Tables
Data collection and refinement statistics.
ctPrp43ΔN•U7•ADP•BeF3- | ctPrp43ΔN•ADP•BeF3-(HR) | ctPrp43ΔN•ADP•BeF3-(LR) | |
|---|---|---|---|
PDBid | 5lta | 5ltj | 5ltk |
Data collection | |||
Space group | P6122 | P212121 | P65 |
Cell dimensions | |||
a, b, c (Å) | 106.39, 106.39, 356.70 | 88.83, 105.64, 119.05 | 184.34, 184.34, 82.32 |
α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 120.0 |
Resolution (Å) | 48.56 – 2.62 (2.70 – 2.62) | 79.02 – 1.78 (1.89 – 1.78) | 92.17 – 3.24 (3.39 – 3.24) |
Rmeas (%) | 5.9 (90.3) | 7.2 (123.5) | 9.8 (76.2) |
I/σ(I) | 22.16 (1.85) | 14.88 (1.64) | 14.65 (2.26) |
CC1/2 (%) | 99.9 (65.2) | 99.9 (61.9) | 99.8 (63.8) |
Completeness (%) | 98.5 (86.8) | 99.6 (98.8) | 98.8 (94.9) |
Redundancy | 5.14 (5.34) | 4.71 (4.66) | 4.00 (3.59) |
Refinement | |||
Resolution (Å) | 48.56 – 2.62 | 67.99 – 1.78 | 92.17 – 3.24 |
No. reflections | 36887 | 107276 | 25304 |
Rwork / Rfree | 19.70/22.97 | 17.67/19.88 | 18.22/21.78 |
No. atoms | |||
Protein | 5605 | 5730 | 5622 |
RNA | 204 | / | / |
Ligand / Ion | 37 | 105 | 42 |
Water | 52 | 688 | 4 |
B-factors(Ų) | |||
Protein | 72.83 | 34.41 | 88.87 |
RNA | 88.65 | / | / |
Ligand / Ion | 55.04 | 47.83 | 83.89 |
Water | 60.51 | 44.29 | 62.19 |
R.m.s. deviations | |||
Bond length (Å) | 0.0028 | 0.0041 | 0.0026 |
Bond angles (°) | 0.78 | 0.87 | 0.67 |
Ramachandran Plot | |||
Favored | 95.98 | 97.34 | 96.28 |
Outlier | 0.0 | 0.0 | 0.0 |
-
Values in parentheses are for the highest resolution shell.
Helicase activity.
nM/min | ± | |
|---|---|---|
ctPrp43 | 0.54 | 0.18 |
ctPrp43 + ctPfa1-GP | 60.03 | 2.75 |
ctPrp43 + ctNtr1-GP | 1.98 | 0.45 |
ctPfa1-GP | 0.46 | 0.28 |
ctPrp43-IDSB + ctPfa1-GP | 12.18 | 0.23 |
ctPrp43-HT+ ctPfa1-GP | 0.68 | 0.16 |
ctPrp43-HL + ctPfa1-GP | 61.87 | 7.14 |
ctPrp43-HT&HL+ ctPfa1-GP | 0.88 | 0.03 |
ATPase activity.
kcat (min−1) | ± | KM (µM) | ± | |
|---|---|---|---|---|
ctPrp43 | 1.81 | 0.03 | 47.23 | 3.40 |
ctPrp43 + U16-RNA | 2.08 | 0.05 | 3.75 | 0.68 |
ctPrp43 + ctNtr1-GP | 4.16 | 0.04 | 5.49 | 0.32 |
ctPrp43 + ctNtr1-GP + U16-RNA | 16.28 | 0.09 | 4.87 | 0.19 |
ctPrp43 + ctPfa1-GP | 36.28 | 0.36 | 8.90 | 0.58 |
ctPrp43 + ctPfa1-GP + U16-RNA | 372.20 | 2.91 | 26.11 | 1.21 |
ctPrp43-IDSB | 5.04 | 0.12 | 16.23 | 2.38 |
ctPrp43-IDSB + ctPfa1-GP | 199.35 | 1.59 | 29.04 | 1.35 |
ctPrp43-IDSB + U16-RNA | 24.99 | 0.37 | 3.42 | 0.38 |
ctPrp43-IDSB + ctPfa1-GP + U16-RNA | 609.38 | 6.15 | 88.14 | 3.50 |
ctPrp43-HT | 13.21 | 0.18 | 91.23 | 5.89 |
ctPrp43-HT + ctPfa1-GP | 60.95 | 0.74 | 13.29 | 1.03 |
ctPrp43-HT + U16-RNA | 10.58 | 0.34 | 53.92 | 8.27 |
ctPrp43-HT + ctPfa1-GP + U16-RNA | 98.61 | 1.84 | 13.19 | 1.52 |
ctPrp43-HL | 6.19 | 0.05 | 14.31 | 0.71 |
ctPrp43-HL + ctPfa1-GP | 86.50 | 0.84 | 14.38 | 0.89 |
ctPrp43-HL + U16-RNA | 6.16 | 0.06 | 5.38 | 0.35 |
ctPrp43-HL + ctPfa1-GP + U16-RNA | 533.55 | 9.53 | 49.49 | 3.86 |
ctPrp43-HT&HL | 7.61 | 0.11 | 16.72 | 1.49 |
ctPrp43-HT&HL + ctPfa1-GP | 113.18 | 1.03 | 25.17 | 1.37 |
ctPrp43-HT&HL + U16-RNA | 5.53 | 0.07 | 5.59 | 0.51 |
ctPrp43-HT&HL + ctPfa1-GP + U16-RNA | 82.28 | 1.31 | 11.88 | 1.18 |





