Coincidence detection and bi-directional transmembrane signaling control a bacterial second messenger receptor
Figures
SEC-MALS reveals a switch of LapD dimers to dimer-of-dimers upon ligand binding.
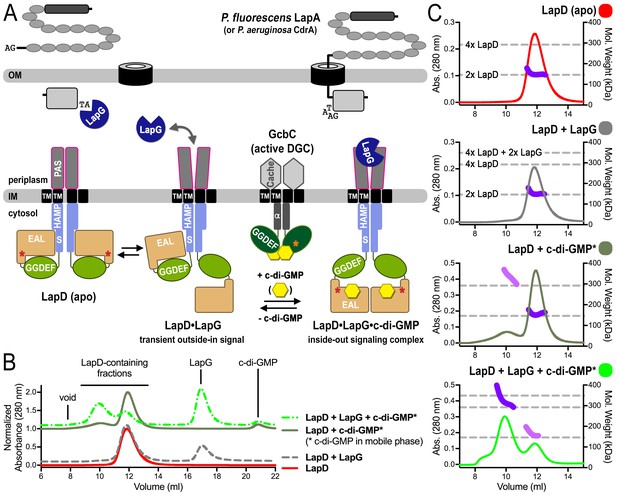
(A) Working model for c-di-GMP-dependent regulation of the periplasmic protease LapG via the inner membrane protein LapD. Concerted conformational changes expose a periplasmic binding site for LapG on LapD, sequestering the protease away from its substrates, the adhesin proteins LapA in P. fluorescens or CdrA in P. aeruginosa. (DGC, diguanylate cyclase; red/orange asterisks indicate interaction helices in LapD/GcbC). (B). Size-exclusion chromatograms for detergent-solubilized LapD in different states. Samples were prepared as described in the Material and Methods. (Asterisks: c-di-GMP was included in the mobile phase). (C) Molecular weight of LapD in solution. Peak fractions were analyzed by in-line SEC-MALS. (Absorbance at 280 nm: Traces colored according to (B); molecular weight determination: Dark and light purple dots; theoretical molecular weights based on sequence: Horizontal dashed lines.) Data are representative of two biological replicates using independent protein preparations.
Mapping of physical interaction motifs onto the crystal structures of LapD and GcbC.
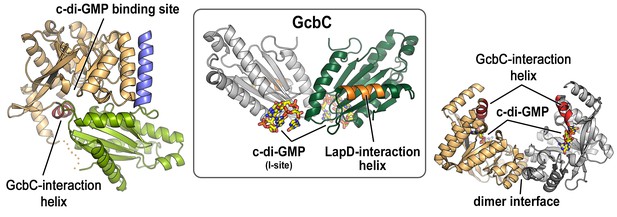
Previous studies identified helical motifs in LapD’s and GcbC’s cytosolic domains (asterisks in Figure 1A) and GcbC’s I-site as features that support interactions between this receptor–DGC pair (Dahlstrom et al., 2015, 2016). Structures: LapD’s S helix-GGDEF-EAL domain, left, PDB code 3pjx; GcbC’s GGDEF domain bound to c-di-GMP, middle, PDB code 5euh; LapD’s EAL domain dimer bound to c-di-GMP, right, PDB code 3pjt).

C-di-GMP binding to LapD results in a 6-fold increase in apparent affinity for LapG.
An equilibrium-saturation binding experiment based on fluorescence anisotropy is shown. See Material and Methods for details. Means and standard errors from two biological and two technical replicates are shown. (Minus c-di-GMP — kD = 11.7 ± 3.8 μM, Bmax = 0.1 ± 0.01; plus c-di-GMP — kD = 1.9 ± 0.3 μM, Bmax = 0.09 ± 0.004.)
SEC-MALS analyses of protein–detergent complexes.
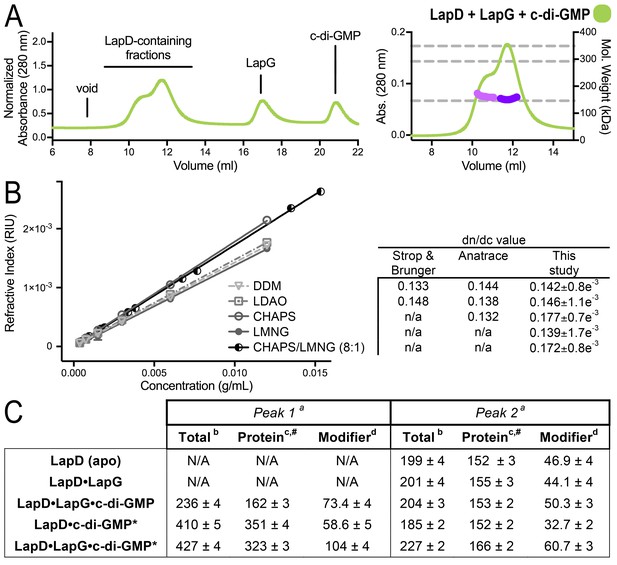
(A) SEC-MALS analysis of LapD incubated with LapG and c-di-GMP. In this experiment, c-di-GMP was omitted from the mobile phase. (B) Experimental determination of detergent dn/dc values. Means and standard errors from three biological replicates are shown. (C) Molecular weights associated with various states of LapD were determined by SEC-MALS. aPeaks 1 and 2 refer to the species eluting at 10–11 min and 12 min, respectively, observed in the associated chromatograms shown in Figure 1C and panel (A) of this figure. bMolecular weight of protein–detergent complex, which is the sum of the protein component molecular weight (c) and the detergent component molecular weight (d). #Theoretical molecular weights: LapD dimer = 146.0 kDa, LapD dimer with one LapG = 168.9 kDa, and LapD dimer of dimers with two LapG molecules = 337.8 kDa. *c-di-GMP present in mobile phase of SEC.
Cysteine crosslinking reveals distinct conformations of apo-, c-di-GMP, and c-di-GMP–LapG-bound LapD.
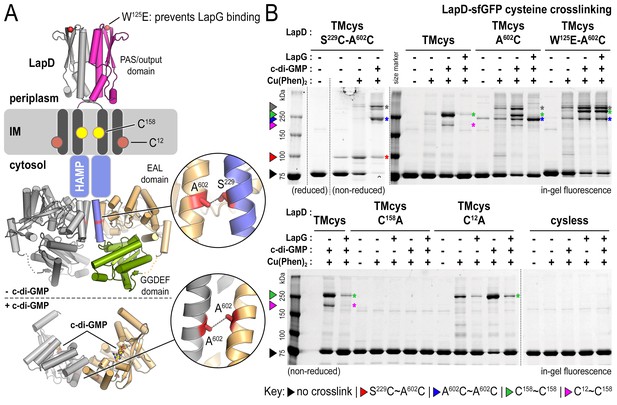
(A) Overview of native and engineered cysteine residues used in crosslinking studies. The composite model of LapD shown is based on the available crystal structures of autoinhibited (top) and c-di-GMP-bound (bottom) domains. (B) Detection of intramolecular and intradimer disulfide bonds. Upper panel: LapDTMcys–sfGFP variants with the indicated cysteine mutation. Colored arrows and asterisks mark specific crosslinking bands. Only the band with grey marking could not be assigned unambiguously to a specific crosslinking sites. Note: The W125E variant of LapD migrates slightly slower than its native counterparts (also seen in Figure 2—figure supplement 1A). Lower panel: LapDTMcys–sfGFP variants with native transmembrane cysteine residues mutated. Experiments with individual protein variants (wild-type and mutants) were repeated at least three times using independent protein preparations. Primary data reproduced here are representative of each replicate.
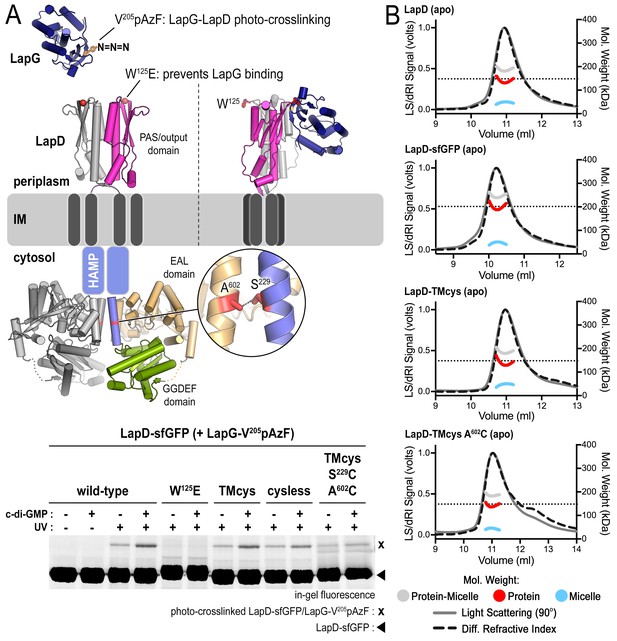
Engineered LapD variants are functional and form constitutive dimers.
(A) LapD cysteine variants TMcys and cysless show similar ability to bind LapG in response to c-di-GMP. Overview of LapG (blue cartoon) binding to LapD’s dimeric output domain (gray and magenta cartoon) in which position V205 of LapG, located at the LapD-binding interface, is replaced with the photo-activatable crosslinking non-canonical amino acid p-azidophenylalanine (pAzF). If LapG is bound to LapD, a covalent crosslink is formed upon UV light exposure, which can then be assessed by SDS-PAGE as a band shift upon SDS-denaturation (without boiling), visualized by in-gel fluorescence (Chatterjee et al., 2014). LapD–sfGFP (native, TMcys, cysless, or TMcys-S229C-A602C) was incubated with LapG-V205pAzF in the absence or presence of c-di-GMP as described in the Materials and methods. The TMcys-S229C-A602C construct is unresponsive to c-di-GMP as it was locked in the autoinhibited state by cysteine crosslinking prior to LapG photocrosslinking to LapD. (B) SEC-MALS data of LapD, LapD–sfGFP, LapD–TMcys, and LapD–TMcys-A602C in their respective apo-states. In the analysis shown here, the TMcys proteins lacked the sfGFP fusion.
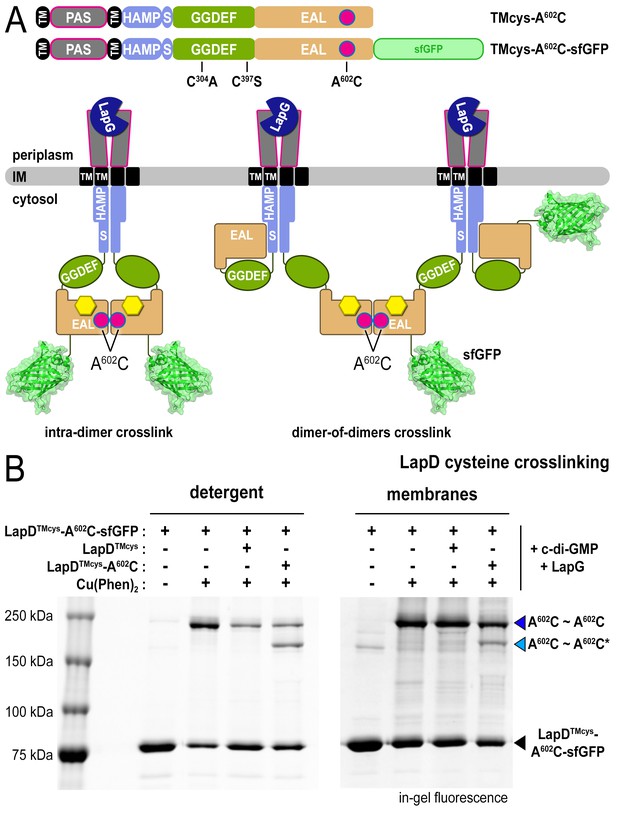
LapD•c-di-GMP•LapG dimer-of-dimers also form in a membrane environment.
(A) Two LapD constructs, one genetically fused to sfGFP and one non-fluorescent, both harboring the A602C mutation, were engineered and expressed separately. In previously proposed models, EAL domain dimerization was thought to occur within a single LapD dimer (left). Under this model, one would only expect a single band shift corresponding to c-di-GMP- and LapG-activated LapD–sfGFP dimers upon oxidation, even if the sfGFP-fused and non-fluorescent variants were present in the membrane. Alternatively, the EAL domains could dimerize across two LapD dimers (right) to form a dimer-of-dimers. Under the latter model, when the two constructs are mixed, activated with c-di-GMP and LapG, oxidized with a disulfide-promoting copper catalyst, denatured in SDS-PAGE, and imaged by in-gel fluorescence, a faster migrating covalent heterodimeric adduct consisting of LapD–sfGFP and LapD (dark) should be observed in addition to the slower migrating complex containing just LapD–sfGFP homodimeric adduct. (B) Complex formation is mediated by dimerization of EAL domains across two LapD dimers rather than within the same LapD dimer. The two LapD variants shown in (A) were expressed separately. Crosslinking via corresponding A602C residues was induced after incubation with c-di-GMP and LapG, either in detergent-solubilized samples (left panel) or upon fusing membrane fractions from the two cultures (right panel). In both detergent and membranes, SDS-PAGE analysis of this crosslinking experiment shows that a heterodimeric adduct containing both LapD–sfGFP and non-fluorescent LapD (lighter blue triangle; asterisk denotes residue from a non-fluorescent LapD) is observed in addition to a species containing only LapD–sfGFP (darker blue triangle). In detergent, non-fluorescent LapD lacking the A602C mutation serves as a competitor, reducing LapD-A602C–sfGFP crosslinking efficiency. Representative data from two independent, biological replicates are shown.
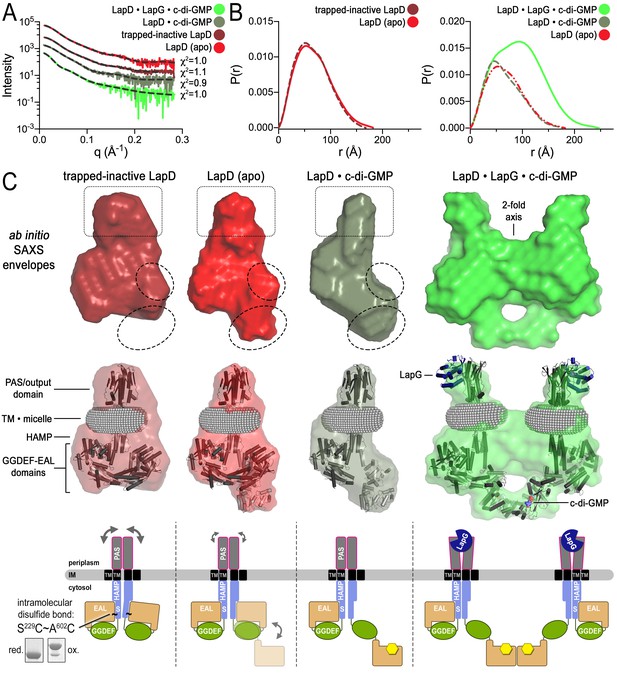
Modeling of SAXS data for distinct LapD states illustrates the conformational changes upon receptor activation.
(A) Primary SAXS data. Solid lines, experimental scattering curves of LapD in the states indicated; dashed lines, theoretical scattering curves of the three-dimensional envelopes shown in panel (C) with χ2 values listed to the right. (B) Real-space pair-wise distance distribution functions for each state of LapD. (C) Modeling of SAXS data. Top: Ab initio three-dimensional envelopes calculated on the basis of the experimental scattering data. Dotted circles and boxes highlight areas of density that change between different states. Middle: Crystal structures of individual domains of LapD docked manually into the envelopes depict interpretations of the ab initio envelope models. Gray spheres represent the detergent corona that surrounds the transmembrane domain. Bottom: Cartoon models of LapD domain movements in each state based on the SAXS data (bottom left inset: SDS-PAGE of purified trapped-inactive LapD used for SAXS analysis). Source files of SAXS data and envelope data are available in Figure 4—source data 2.
-
Figure 4—source data 1
Statistics associated with the analysis of LapD small-angle X-ray scattering data.
All analyses were performed using the indicated programs included in the ATSAS 2.7.1 software package (Petoukhov et al., 2012). Rg, radius of gyration; Dmax, maximal particle dimension; Porod volume, volume of scattering particle; NSD, normalized spatial discrepancy; Rflex and Rsigma, as previously defined (Tria et al., 2015).
- https://doi.org/10.7554/eLife.21848.010
-
Figure 4—source data 2
SAXS data and envelope model files related to Figure 4.
The zip archive contains buffer-subtracted scattering raw data (folder ‘raw_data’, extension ‘.dat’) and GNOM files (folder ‘gnom_output’, extension ‘.out’) that contain original data, real space distance distribution functions, and associated statistics. The folder ‘damfilt_pdb’ contains the final files (extension ‘.pdb’) of the envelope modeling.
- https://doi.org/10.7554/eLife.21848.011
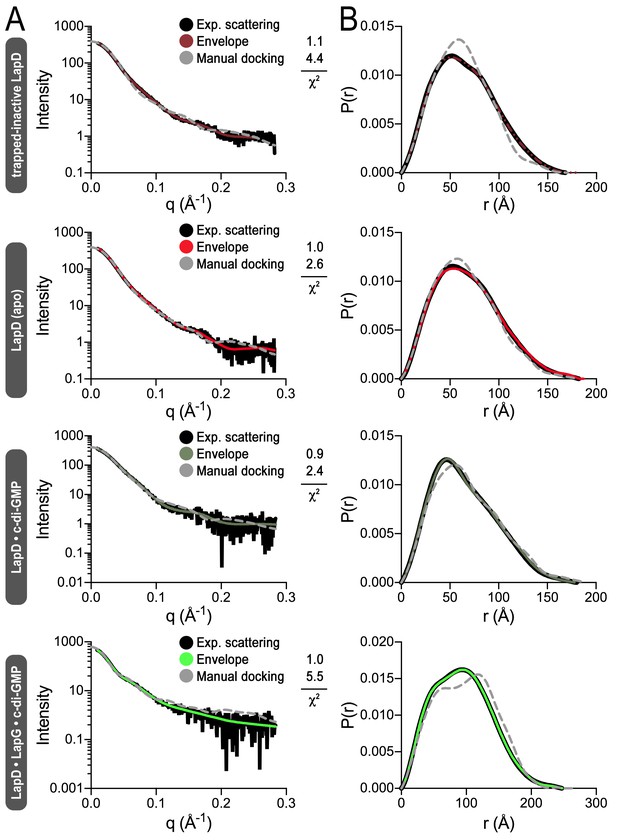
Fitting of various models to LapD small-angle X-ray scattering data.
(A) Experimental scattering data for the indicated states of LapD (black lines) overlayed with theoretical scattering curves of models generated from envelopes (solid, colored lines) and manual dockings (dashed, grey lines) shown in Figure 4B, with their associated χ2 fit values. (B) Real-space distance distribution functions for these theoretical models of LapD overlayed on the experimentally derived distribution function (black).
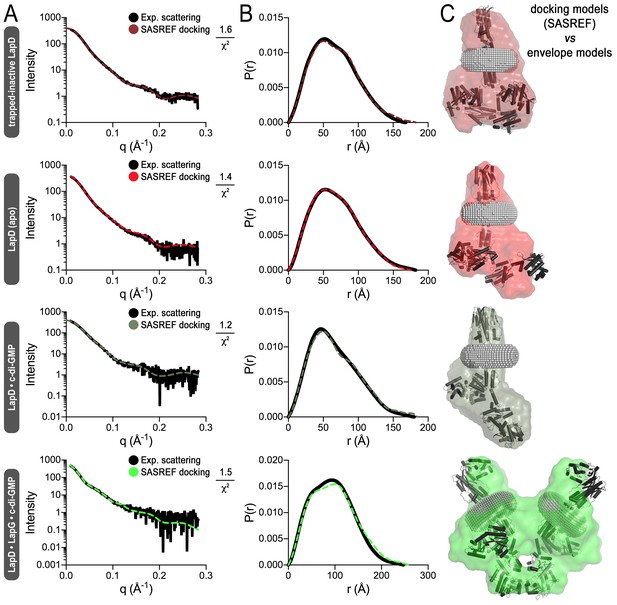
Simulated annealing and rigid-body fitting of crystal structures of individual LapD domains to full-length LapD small-angle X-ray scattering data.
(A) Experimental scattering data of the indicated states of LapD (black lines) overlayed with theoretical scattering curves of these SASREF-generated models (dashed, colored lines), with their associated χ2 fit values. (B) Real-space distance distribution functions for these theoretical models of LapD overlayed on the experimentally derived distribution function (black). (C) Docking of the SASREF models for each LapD state into their respective three-dimensional envelopes shows reasonable agreement between the two independent analyses of the SAXS scattering data.
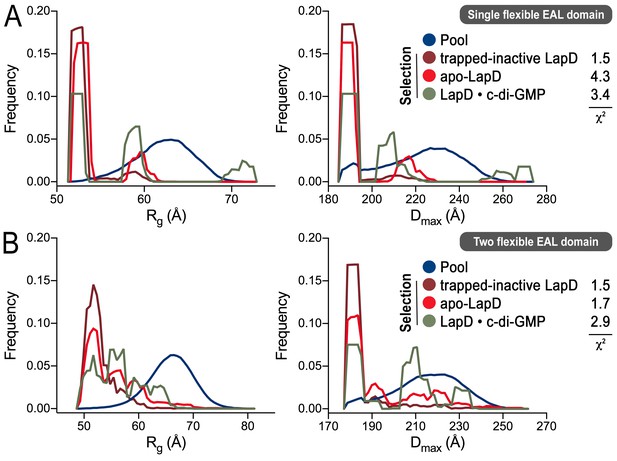
Ensemble optimization method (EOM) analysis of the LapD SAXS data.
On the basis of the experimental SAXS data for three states (trapped-inactive, apo, and c-di-GMP-bound), ensembles of LapD conformations were selected from a random pool of 10,000 models with either one or two flexible EAL domains in a LapD dimer. While the χ2 fit values indicate imperfect modeling of the SAXS data by this approach (probably due to the constraints that we imposed on the global LapD conformation), there is a clear trend in selections for the three different states with regard to the Rg (left panels) and Dmax (right panels) distributions of the selected conformers, consistent with other experiments and analyses described in this report. (A) Selection from a pool generated with one EAL domain of a LapD dimer being flexibly linked to the GGDEF domain. (B) Selection from a pool generated with both EAL domains of a LapD dimer being flexibly linked to the GGDEF domain. See Material and methods for details and Figure 4—source data 1 for associated statistics.
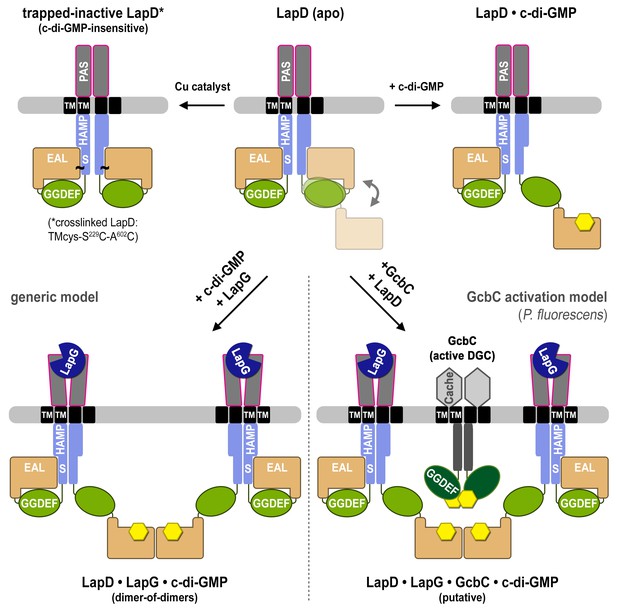
Proposed mechanism for activation of the LapD receptor.
In the absence of c-di-GMP, apo LapD (top center) adopts a dynamic ‘half-open’ state, transiently exposing the nucleotide-binding site of one EAL domain, which can be occluded by locking the receptor in a c-di-GMP-insensitive autoinhibited state (top left; “trapped-inactive") by covalently crosslinking the S helix to the EAL domain (S229C ~ A602C). Upon binding of c-di-GMP to the native receptor’s accessible EAL domain, LapD adopts a more extended state (top right). Only when LapG and c-di-GMP bind the receptor together does LapD form dimer-of-dimers, for which the interface is mediated by EAL domain dimerization across two receptor dimers (bottom left). On the basis of recent findings that the diguanylate cyclase GcbC interacts with and specifically activates LapD (Dahlstrom et al., 2015, 2016), we hypothesize that activated LapD dimer-of-dimers may be stabilized by GcbC (bottom right) to ensure prolonged, high-affinity sequestration of LapG in the periplasm.





