Concerted action of the MutLβ heterodimer and Mer3 helicase regulates the global extent of meiotic gene conversion
Figures
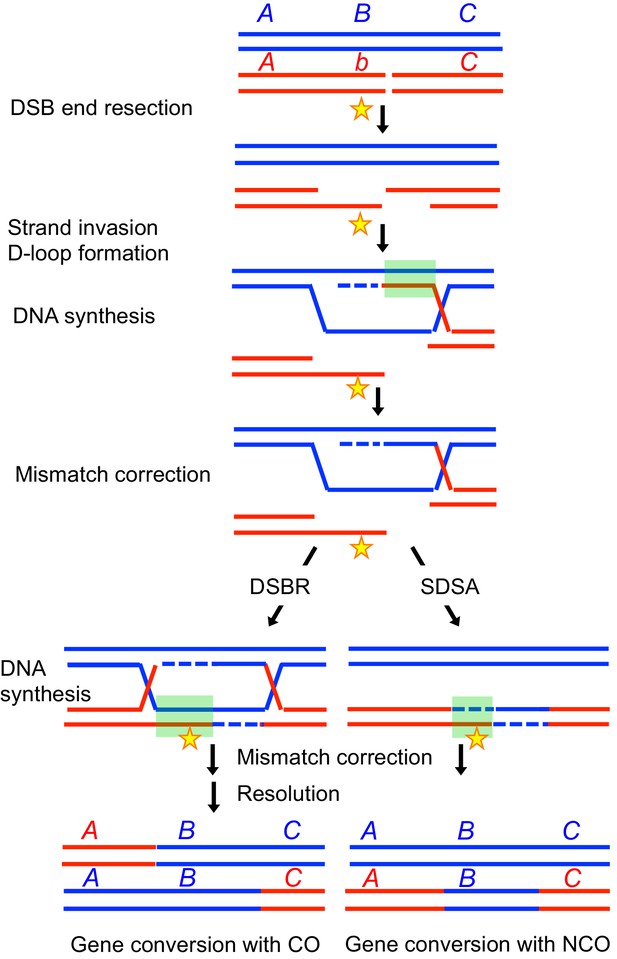
Effect of gene conversion during meiotic recombination on allele shuffling and erosion of cis-acting hotspot sequences.
Two homologous chromatids are shown in red and blue, the red one having a DSB formed by Spo11. A, B and C represent alleles, with the b allele being present on the red homolog. The star represents a cis-acting hotspot promoting sequence. Following DNA end resection and strand invasion of the intact DNA duplex, the red homolog sequences are copied from the blue one, creating heteroduplexes (indicated by a green square), that are next corrected by mismatch repair, leading either to gene conversion or restoration. After gene conversion, the b allele on the broken chromosome has been converted to the B allele, and the hotspot promoting sequence has been converted to the blue sequence.
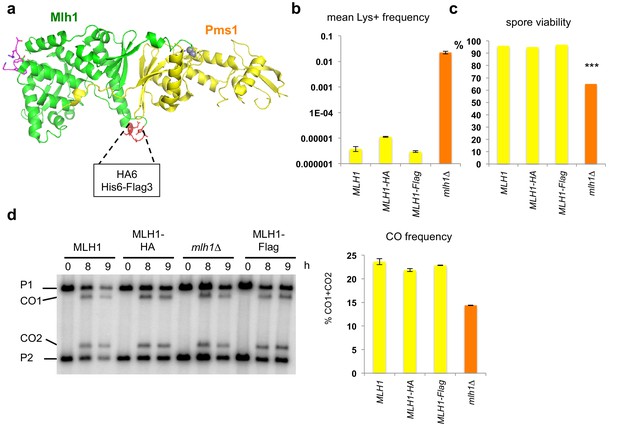
Mlh1 alleles tagged internally in the Cter domain are fully functional for MMR and meiotic recombination.
(a) Crystal structure of the C-terminal region of S. cerevisiae Mlh1-Pms1 heterodimer (pdb code 4FM0) (Gueneau et al., 2013). The Mlh1 and Pms1 regions are colored in green and yellow, respectively. The two metal ions of the endonuclease site are represented by grey spheres. The peptide containing the Mlh1-binding motif for Exo1 is colored in magenta. (b) Mutator assay. Frequency of reversion to Lys+ in cells containing the indicated MLH1 genotype at its endogenous locus. Values are the mean of 9 independent colonies ± SEM. (c) Spore viability of diploid SK1 strains bearing the indicated MLH1 genotype at its endogenous locus. MLH1: VBD1311 (188 tetrads), Mlh1-HA: VBD1456 (22 tetrads), Mlh1-Flag: VBD1337 (26 tetrads), mlh1∆: VBD1494 (178 tetrads). ***p<5.10−5, Fisher’s exact test. (d) Crossing over frequency at the HIS4LEU2 hotspot monitored by Southern blot at the indicated times in meiosis. Positions of parental bands (P1 and P2) and of the recombinant crossover products (CO1 and CO2) are indicated. Graph shows quantification at 9 hr from two independent biological replicates ± SEM. Same strains as in (c).
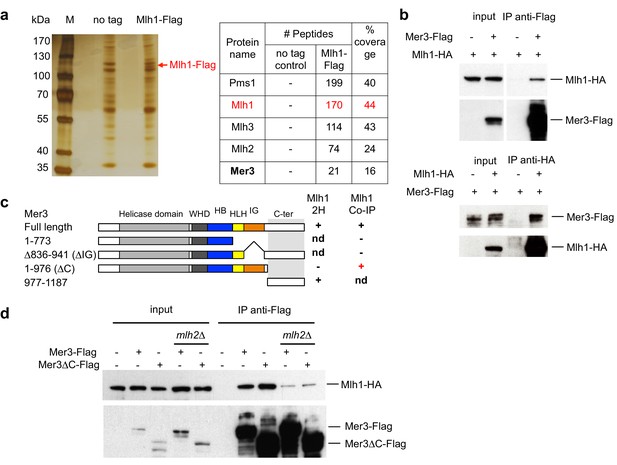
Mlh1 interacts with the meiosis-specific Mer3 helicase.
(a) Affinity pull-down of Mlh1-Flag from cells at 4 hr in meiosis. M: marker, No tag: VBD1311, Mlh1-Flag: VBD1337. Left: silver-stained gel of pulled-down proteins. Right: mass-spectrometry analysis of the most significant proteins pulled-down with Mlh1. One representative experiment is shown. (b) Reciprocal co-immunoprecipitation between Mer3-Flag and Mlh1-HA from meiotic cells at 4 hr in meiosis, analyzed by Western blot. Mer3-Flag: VBD1420, Mlh1-HA: VBD1456, Mer3-Flag Mlh1-HA: VBD1454. (c) Comparative analysis of two-hybrid interactions and co-IP in meiotic cells. The domain limits are based on Mer3 modeling with the Mer3 family-related Brr2 helicase structure (Santos et al., 2012). nd: not determined. (d) Co-IP between Mer3-Flag and Mlh1-HA at 4 hr in meiosis, in the presence or absence of MutLβ component Mlh2. Mlh1-HA: VBD1456, Mer3-Flag Mlh1-HA: VBD1454, Mer3∆C-Flag Mlh1-HA: VBD1490, Mer3-Flag Mlh1-HA mlh2∆: VBD1550, Mer3∆C-Flag Mlh1-HA mlh2∆: VBD1552.
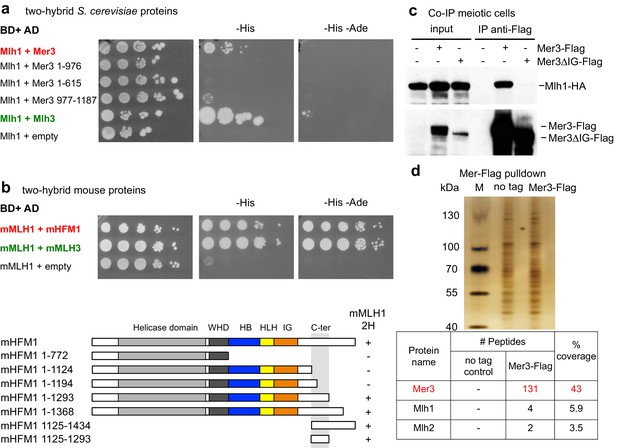
Interaction between yeast or mouse Mer3 and Mlh1 and pulldown of Mlh1 and Mlh2 by Mer3.
Serial dilutions of strains expressing the different fusion proteins were plated on minimal media lacking the indicated aminoacids to select for interactions. (a) Interaction between the S. cerevisiae proteins or their domains with the indicated coordinates. Mlh1 and Mlh3 interaction is used as a positive control. (b) Interaction between the mouse proteins. mMLH1 and mMLH3 interaction is used as a positive control. Below: diagram showing the tested mHFM1 domains for their interaction with mMLH1. Domains are color-coded as in Figure 2c. (c) Co-IP between Mer3-Flag and Mlh1-HA from meiotic cells at 4 hr in meiosis, analyzed by Western blot. Mlh1-HA: VBD1564, Mer3-Flag Mlh1-HA: VBD1576, Mer3∆IG-Flag Mlh1-HA: VBD1579. (d) Affinity pull-down of Mer3-Flag from cells at 4 hr in meiosis. No tag: VBD1311, Mer3-Flag: VBD1420. Top: silver-stained gel of pulled-down proteins. Bottom: mass-spectrometry analysis of the MutL proteins pulled-down with Mer3. One representative experiment is shown.
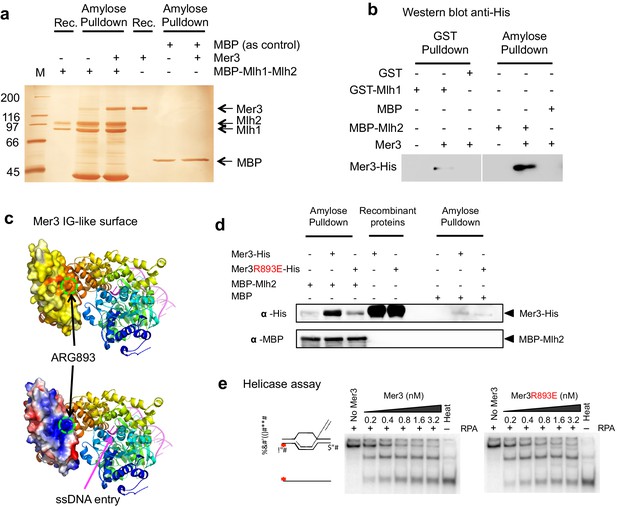
Mer3 interacts directly with the Mlh1-Mlh2 (MutLβ) heterodimer.
(a) Silver-stained gel showing the direct interaction between purified Mer3 and Mlh1-Mlh2 (MutLβ) complex tagged with MBP on Mlh2. MBP-tagged MutLβ or MBP were bound to amylose resin and incubated with Mer3-His in the presence of 80 mM NaCl. Proteins were eluted with maltose. (b) Western Blot analysis using anti His antibody showing the pulldown of purified Mer3-His by GST-Mlh1 or by MBP-Mlh2 in the presence of 150 mM NaCl. Both panels are from the same exposure time of the same membrane. (c) Structural model of the IG-like domain of Mer3, based on Brr2 helicase structure (Santos et al., 2012). Top diagram: surface of the IG-like domain showing aminoacid conservation, from low (white) to high (red). Bottom: Electrostatic potential indicated by a color code, from positive (blue) to negative (red) charge. The position of Arg893 is indicated. (d) Western Blot showing the pulldown of purified Mer3-His or Mer3R893E-His by MBP-Mlh2 or MBP alone as a control. Pulldown was done in the presence of 150 mM NaCl. (e) Helicase assays with Mer3 or Mer3R893E on labeled D-loop substrate. 'Heat’, heat-denatured DNA substrate indicates the position of ssDNA. Assays were performed with or without RPA (20 nM) as indicated.
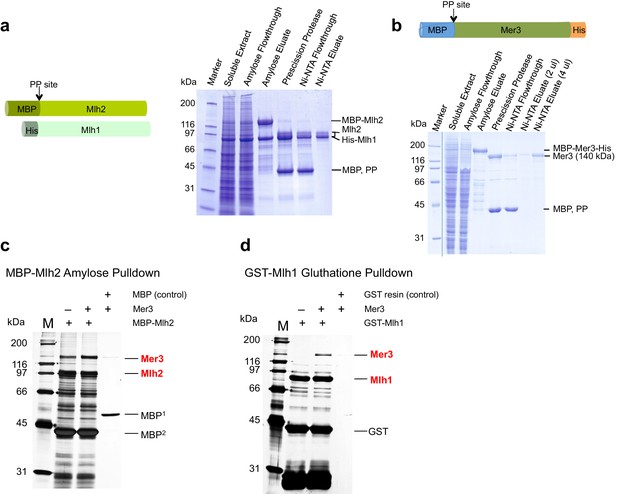
Purification of yeast Mlh1-Mlh2 and interaction with purified Mer3.
(a) Scheme of Mlh2 and Mlh1 expression constructs. MBP, Maltose binding protein tag; His, 8x histidine tag; PP site, PreScission Protease cleavage site. Right: 4% to 15% SDS-PAGE showing samples from a representative purification of Mlh1-Mlh2. The MBP tag was cleaved by using PreScission protease. We observed a single band for Mlh1, which migrated at a position corresponding to its molecular weight of 87 kDa. The 78 kDa-large Mlh2 polypeptide co-migrated with Mlh1, with a fraction of the protein (~20%) migrating above it. (b) Scheme of Mer3 expression construct. MBP, Maltose binding protein tag; His, 10x histidine tag. A 7.5% SDS-PAGE showing various fractions from Mer3 purification. The MBP tag was cleaved by using PreScission protease. (c) Silver-stained gel showing the direct interaction between purified Mer3 and Mlh2. MBP-Mlh2 or MBP alone was incubated with Mer3-His in the presence of 80 mM NaCl. MBP1 is expressed and purified from E.coli and MBP2 results from the cleavage of MBP tag from Mlh2 protein. (d) Silver-stained gel showing the direct interaction between purified Mer3 and Mlh1. GST-Mlh1 or GST resin was incubated with Mer3-His in the presence of 80 mM NaCl.
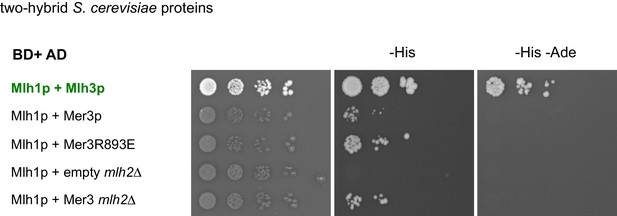
Two hybrid interaction between Mer3 and Mlh1 is independent of Mlh2.
Serial dilutions of strains expressing the different fusion proteins were plated on minimal media lacking the indicated aminoacids to select for interactions.
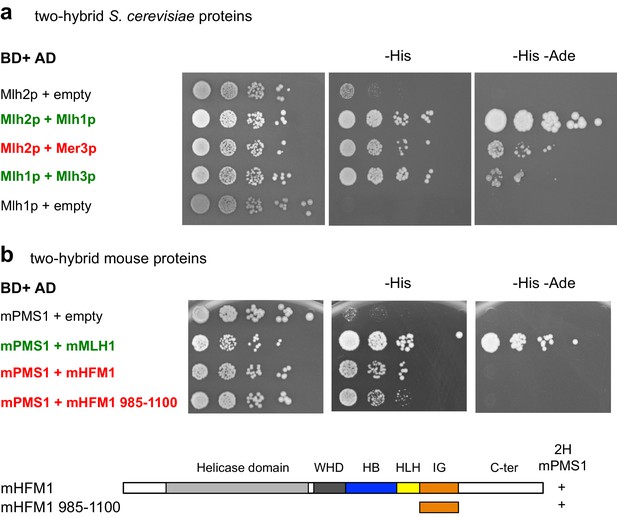
Mer3 and Mlh2 interact in yeast two-hybrid assays.
(a) and (b): Serial dilutions of strains expressing the different fusion proteins were plated on minimal media lacking the indicated aminoacids to select for interactions. (a) Interaction between the S. cerevisiae proteins. Mlh1 and Mlh2 interaction is used as a positive control. (b) Interaction between the mouse proteins or their domains with the indicated coordinates. mMLH1 and mPMS1 interaction is used as a positive control. Below: diagram showing the tested mHFM1 domain for its interaction with mPMS1. Domains are color-coded as in Figure 1b.
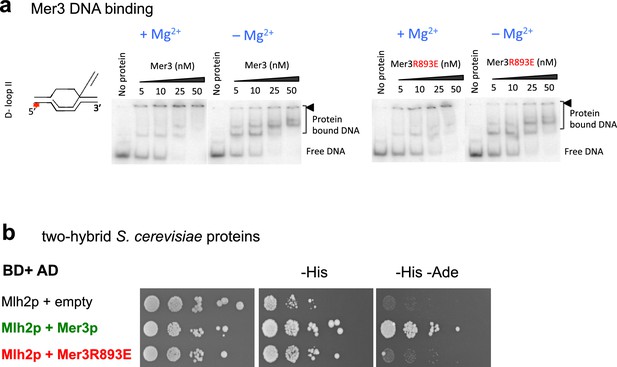
The Mer3R893E mutation disrupts interaction with Mlh2 but not its DNA binding.
(a) Mer3 or Mer3R893E EMSA were performed using an oligonucleotide based D-loop DNA substrate with either 2 mM magnesium (+Mg2+) or 3 mM EDTA (–Mg2+). The black triangle indicates the position of gel wells. (b) Yeast two hybrid interaction. Serial dilutions of strains expressing the different fusion proteins were plated on minimal media lacking the indicated aminoacids to select for interactions.
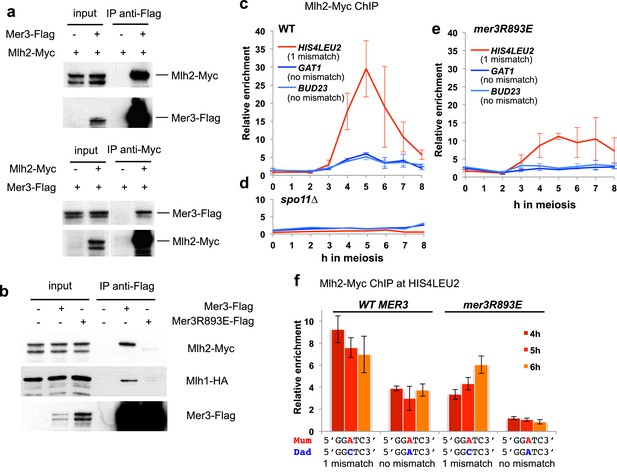
Mlh2 binds meiotic recombination intermediates.
(a) Reciprocal co-IP between Mer3-Flag and Mlh2-Myc from meiotic cells at 4 hr in meiosis, analyzed by Western blot. Mer3-Flag: VBD1420, Mlh2-Myc: VBD1628, Mer3-Flag Mlh2-Myc: VBD1670. (b) Co-IP between Mer3- or Mer3R893E-Flag and Mlh2-Myc or Mlh1-HA from meiotic cells at 4 hr in meiosis, analyzed by Western blot. Mlh1-HA Mlh2-Myc: VBD1630; Mer3-Flag Mlh1-HA Mlh2-Myc: VBD1629; Mer3R893E-Flag Mlh1-HA Mlh2-Myc: VBD1681. (c) Mlh2-Myc levels at three meiotic recombination hotspots assessed by ChIP and qPCR at the indicated times during a meiotic time-course (VBD1670). (d) Same as in (c) but in a DSB-deficient spo11∆ strain (VBD1702). (e) Same as in (c) but in the mer3R893E strain (VBD1637). (f) Effect of polymorphism at the HIS4LEU2 hotspot on Mlh2 binding. Mlh2-Myc association with the HIS4LEU2 hotspot was assessed by ChIP at the indicated times in meiosis in strains containing either the wild-type or R893E mer3 allele, in strains with one base mismatch or without a mismatch at the HIS4LEU2 hotspot. WT one base mismatch: VBD1670; WT no mismatch: VBD1710; mer3R893E one base mismatch: VBD1637; mer3R893E no mismatch: VBD1706. (c), (e) and (f): Values are the mean ± SEM from two independent experiments.
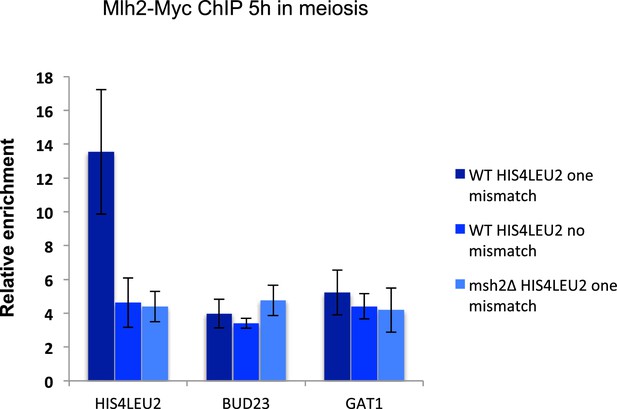
Polymorphism at the HIS4LEU2 hotspot triggers Mlh2 recruitment by Msh2.
Mlh2-Myc association with the HIS4LEU2, BUD23 and GAT1 hotspots was assessed by ChIP at 5 hr in meiosis in strains with the indicated genotype. WT HIS4LEU2 one mismatch: VBD1628, WT HIS4LEU2 no mismatch: VBD1707, msh2∆ HIS4LEU2 one mismatch: VBD1704. Values are the mean and error bars indicate SEM from two independent experiments.
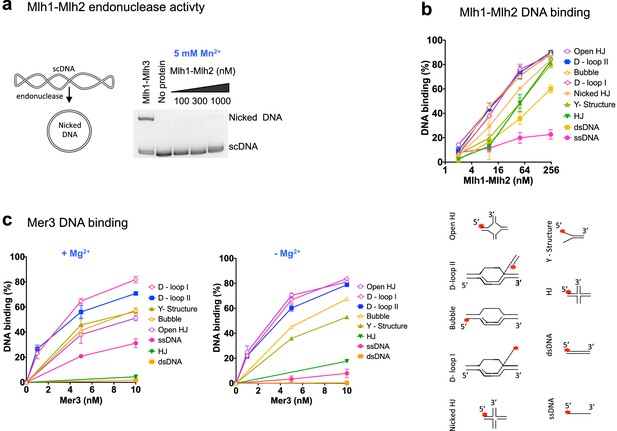
Recombinant Mer3 and MutLβ preferentially bind DNA substrates mimicking early recombination intermediates.
(a) Nuclease assay was performed with Mlh1-Mlh2 (as indicated) or Mlh1-Mlh3 (300 nM) on the indicated super-coiled circular plasmid DNA substrate (scDNA). Mlh1-Mlh3 is shown as a positive control producing nicked circular DNA (b) Quantitation of electrophoretic mobility shift assays with Mlh1-Mlh2 and various oligonucleotide-based DNA substrates in the presence of magnesium (2 mM). Values are the mean ± SEM from two independent experiments. (c) Electrophoretic mobility shift assays with Mer3 and the indicated DNA substrates, in the presence of either 2 mM magnesium (+Mg2+) or 3 mM EDTA (–Mg2+). Values are the mean ± SEM from two to tree independent experiments.
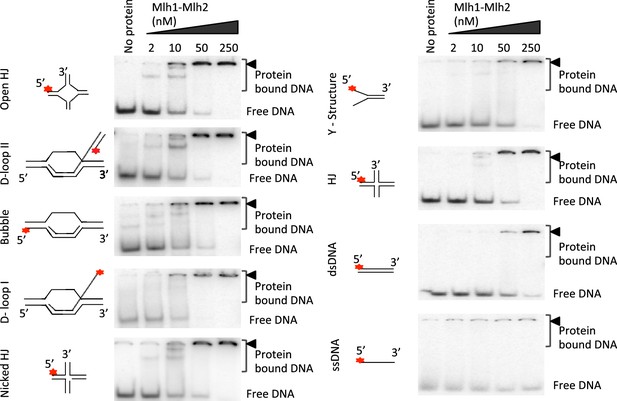
DNA binding specificities of Mlh1-Mlh2.
Representative experiments from Figure 6b, analyzed by 6% PAGE.
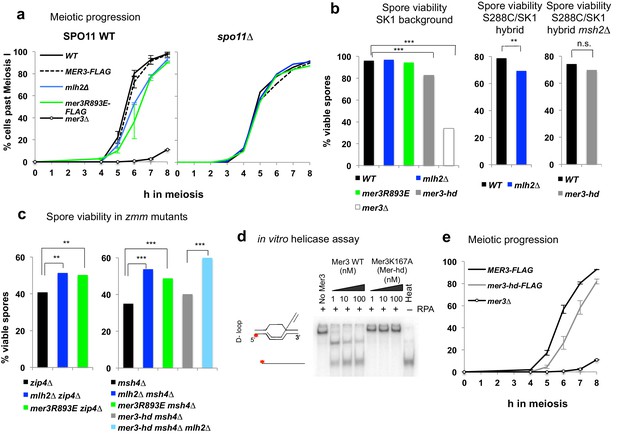
Disruption of the Mlh2/Mer3 interaction improves spore viability of zmm mutants.
(a) Meiotic progression as assessed by DAPI staining of strains with the indicated genotype: WT (VBD1311), Mer3-Flag (VBD1420), mlh2∆ (VBD1631), Mer3R893E-Flag (VBD1635), mer3∆ (VBD1414), spo11∆ (VBD1382), spo11∆ Mer3-Flag (VBD1794), spo11∆ mlh2∆ (VBD1796), spo11∆ Mer3R893E-Flag (VBD1795). (b,c) Spore viability assays of strains with the indicated genotype. Fisher’s exact test, **p<5.10−3; ***p<5.10−5; n.s.: p>0.05. Numbers of tetrads dissected and strains names are in Supplementary file 1. (d) Helicase assays with recombinant Mer3 or Mer3K167A in the presence of 20 nM RPA on a D-loop substrate. 'Heat’, heat-denatured DNA substrate indicates the position of ssDNA. (e) Meiotic progression as assessed by DAPI staining of strains with the indicated genotype: Mer3-Flag (VBD1420), Mer3-hd-Flag (VBD1750) or mer3∆ (VBD1414). (a), left panel and (e): Values are the mean ± SEM from two independent experiments.
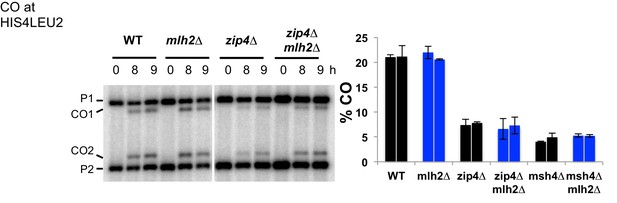
Effect of the Mer3-Mlh2 interaction on CO frequency at the HIS4LEU2 hotspot.
Crossing over frequency at the HIS4LEU2 hotspot monitored by Southern blot at the indicated times in meiosis. The positions of parental bands (P1 and P2) and of the recombinant crossover products (CO1 and CO2) are indicated. Graph shows quantification from two independent biological replicates ± SEM. WT: VBD1311, mlh2∆: VBD1631, zip4∆: VBD1082, zip4∆ mlh2∆: VBD1602, msh4∆: VBD1676, msh4∆ mlh2∆: VBD1682.
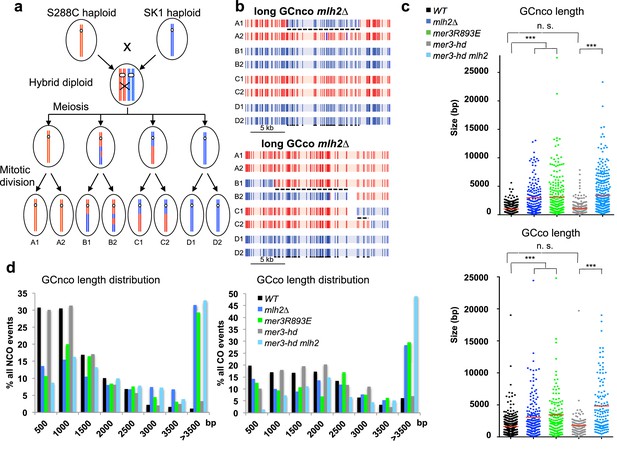
The Mlh2/Mer3 interaction limits the extent of gene conversions genome-wide.
(a) Scheme of the experimental system to measure genome-wide recombination events. After meiosis, the four haploid spores were allowed to perform one mitosis and micromanipulated, in order to sequence DNA of the two daughters, allowing the recovery of the 8 DNA recombined strands from the initial diploid cell. (b) Blow up of NCO and CO events of a mlh2∆ meiosis, showing very long hDNA tracts (referred to as GC, or gene conversions) more frequent in this mutant. The 8 strands are shown. Each vertical bar represents a nucleotide polymorphism between the two strains (blue: of SK1 origin; red: of S288C origin). (c) Gene conversion lengths among NCO and CO events of meioses from the indicated genotype. The horizontal bar indicates the mean value of all events from the meioses (4 meioses of WT, and 2 meioses of each other relevant genotype). Wilcoxon rank sum test, ***p<5.10−5. n.s.: p>0.05 (d) Distribution of gene conversion lengths among NCO and CO events of meioses each from the indicated genotype.
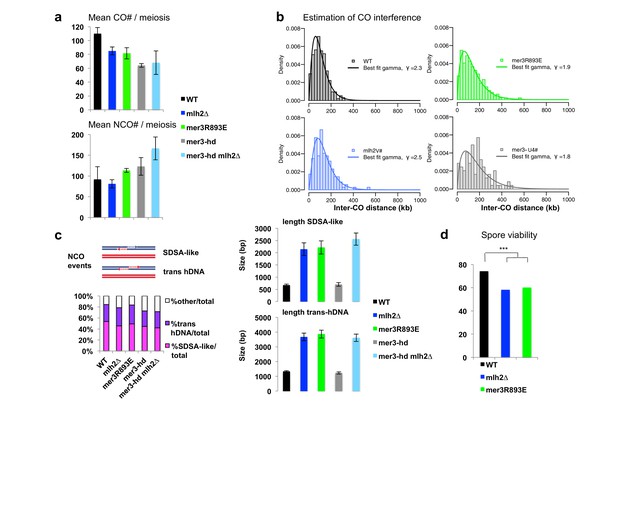
CO and NCO numbers and other events in the hybrid strains.
(a) CO and NCO numbers per meiosis. Numbers are the mean number ± S.E. per meiosis for each indicated genotype. (b) Estimation of crossover interference in the hybrids. For each strain, the density of experimental inter-CO distance distribution (histogram with bin size of 25 kb) and best fit gamma distribution (solid line) is shown. The shape parameter γ of the fit is given in the legend box, and is indicative of the strength of crossover interference (γ = 1: no interference, γ > 1: positive interference). Although all four strains exhibit positive interference, their inter-CO distance distribution is significantly different in pairwise comparison (p<0.05, Kolmogorov-Smirnov test) except for mlh2∆ and mer3R893 which are not significantly different from each other. (c) hDNA associated with NCO events. The two main hDNA categories, SDSA-like and trans hDNA are cartooned. Left panel: distribution of the different types of hDNA in the hybrids. Right panels: hDNA lengths of the two types of hDNA in the hybrids. Values are the mean±S.E.M. (d) Spore viability assays of the msh2∆ hybrid strains with the indicated genotype. Fisher’s exact test, **p<5.10−3. Numbers of tetrads dissected and strains names are in Supplementary file 1.
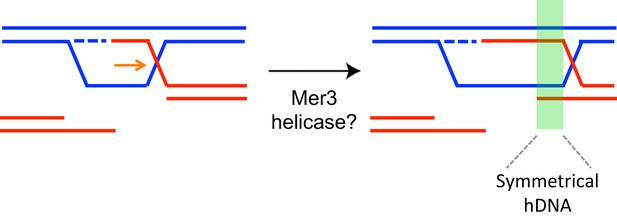
Formation of symmetrical heteroduplex upon D-loop extension in the direction opposed to DNA synthesis.
The region of resulting symmetrical heteroduplex is indicated by a green rectangle. It contains two duplexes each with 2 strands of different parents.
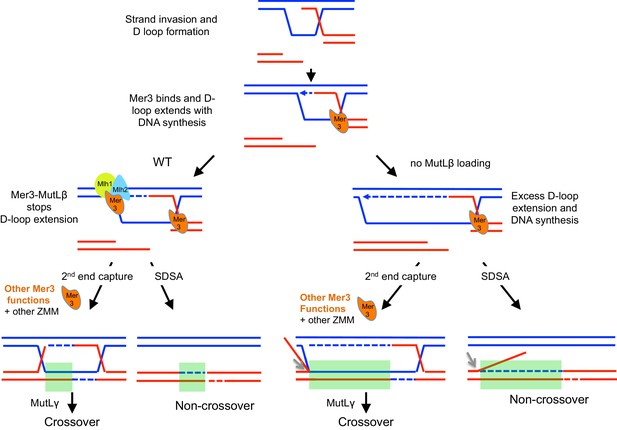
Model for the actions of Mer3 and MutLβ on recombination sites.
Following DSB formation and strand invasion, Mer3 (in orange) binds the resulting D-loop and DNA synthesis begins (dotted blue arrow). MutLβ (light blue and yellow complex) interaction with Mer3 then acts as a lock to stop D-loop extension in DNA synthesis direction (left panel). In the absence of Mlh2, D-loop will extend significantly further in the DNA synthesis direction (right panel). After this step, Mer3 is expected to have other, procrossover functions, partly dependent on its helicase activity, acting together with the other ZMM proteins and MutLγ. Grey arrows indicate that endonuclease activity should remove the flap generated by overextension of the D-loop in the absence of MutLβ. The light green rectangles indicate the gene conversion tracts, longer in the absence of MutLβ.
Additional files
-
Suplementary file 1
Spore viabilities.
- https://doi.org/10.7554/eLife.21900.021
-
Supplementary file 2
Genotypes of strains used in this study.
- https://doi.org/10.7554/eLife.21900.022
-
Supplementary file 3
Strains used in each Figure panel.
- https://doi.org/10.7554/eLife.21900.023
-
Supplementary file 4
Primary data for graphs in each figure.
- https://doi.org/10.7554/eLife.21900.024





