Molecular architecture of the 90S small subunit pre-ribosome
Figures
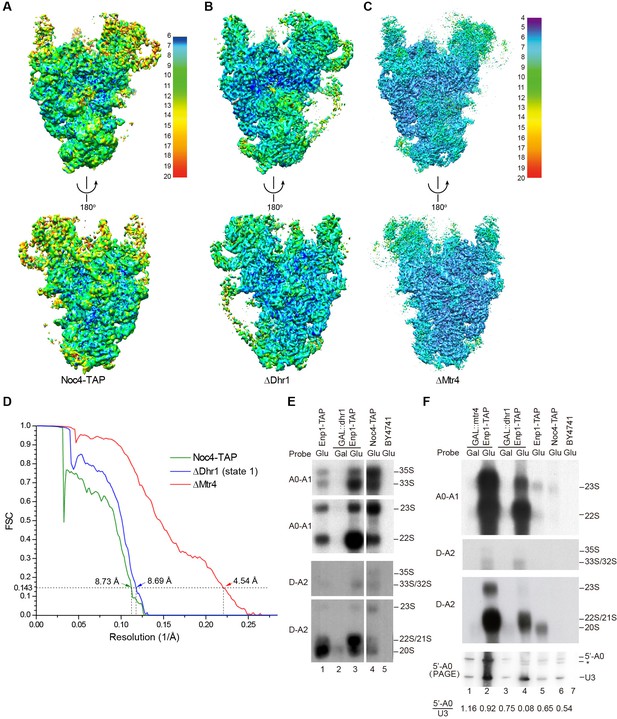
Cryo-EM analysis of 90S.
(A–C) Local resolution cryo-EM maps of 90S derived from the Noc4-TAP (A), ΔDhr1 (state 1) (B) and ΔMtr4 (C) samples. The maps in A and B are colored with the same scale. Each map is shown in front and back views. (D) Gold-standard FSC curves for three constructions. (E–F) Northern blot analysis of RNA in purified pre-ribosomes. The GAL::dhr1/Enp1-TAP and GAL::mtr4/Enp1-TAP strains were grown in YPG (Gal) and shifted to YPD (Glu) for 14 hr to deplete the helicases. Pre-ribosomes were immunoprecipitated from equal OD260 units of cell lysates and extracted for RNA. RNA was resolved in 1.2% agarose-formaldehyde gels or 8% polyacrylamide-8M urea gels (PAGE) and hybridized to 32P-labeled probes indicated. The wild-type cells have a much lower yield of pre-ribosomes as compared to the helicase-depleted cells. Two experiments are shown in E and F. The 35S and 33S RNAs were not efficiently transferred to membranes in F. Asterisk indicates a degradation product of the 5'-A0 fragment. The volume ratios of 5'-A0 fragment to U3 are calculated.
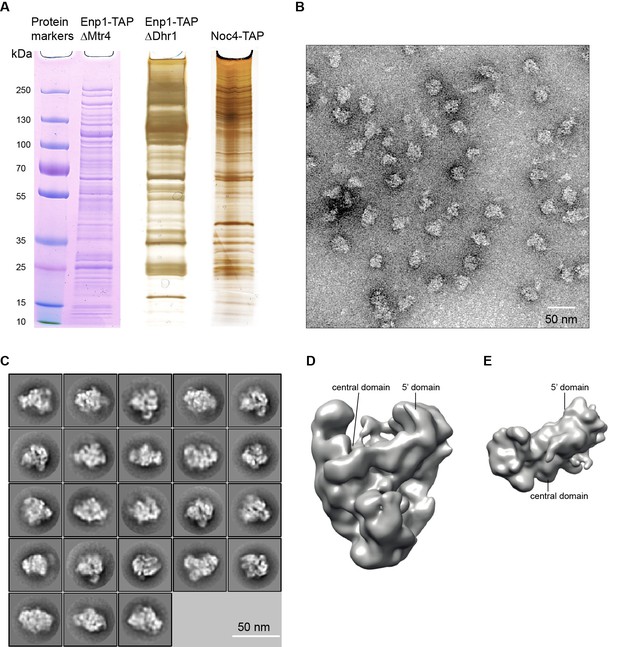
Negative stain EM analysis of 90S samples.
(A) SDS-PAGE of the ΔMtr4/Enp1-TAP, ΔDhr1/Enp1-TAP and Noc4-TAP samples. The first sample was stained by Coomassie-blue and the latter two were silver-stained. (B) Electron micrograph of negative stain Noc4-TAP particles. Bar = 50 nm. (C) Representative reference-free 2D class averages of negative stain Noc4-TAP particles. Bar = 50 nm. (D) Refined EM density map of negative stain Noc4-TAP particles. (E) The 40S ribosome structure was used as the initial model for 3D classification. The initial model is shown in the original orientation relative to the 90S map in D. Note that the 5' domain and the central domain have similar positions in the two maps.
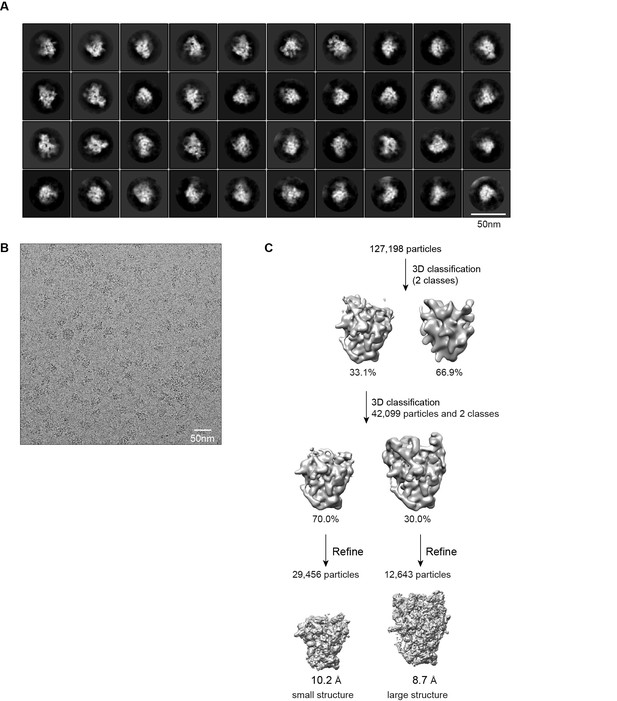
Cryo-EM analysis of Noc4-TAP 90S samples.
(A) Representative reference-free 2D class averages of Noc4-TAP particles. Bar = 50 nm. (B) Electron micrograph of Noc4-TAP particles. Bar = 50 nm. (C) Flowchart of 3D classification and refinement of Noc4-TAP particles.
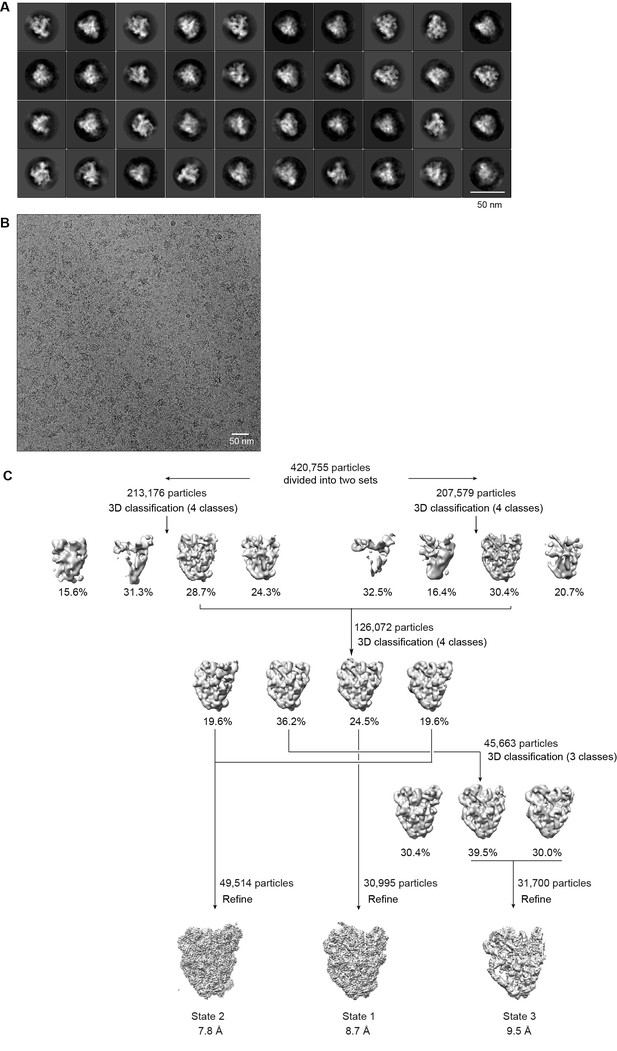
Cryo-EM analysis of ΔDhr1 90S samples.
(A) Representative reference-free 2D class averages of ΔDhr1 90S particles. Bar = 50 nm. (B) Electron micrograph of ΔDhr1 90S particles. Bar = 50 nm. (C) Flowchart of 3D classification and refinement of ΔDhr1 90S particles.
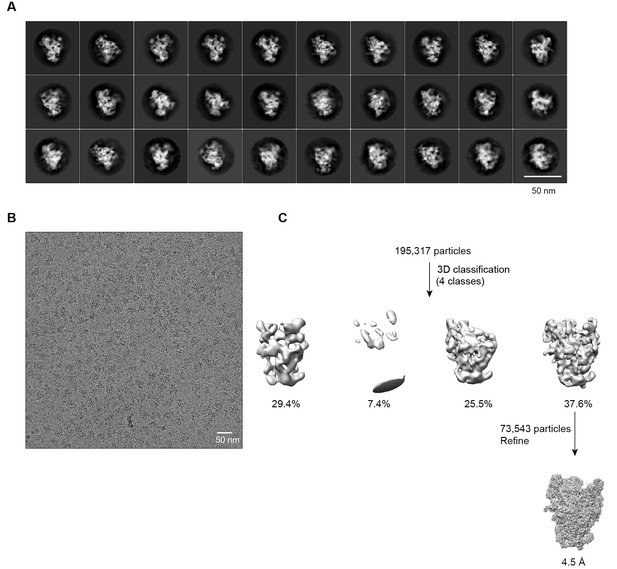
Cryo-EM analysis of ΔMtr4 90S samples.
(A) Representative reference-free 2D class averages of ΔMtr4 90S particles. Bar = 50 nm. (B) Electron micrograph of ΔMtr4 90S particles. Bar = 50 nm. (C) Flowchart of 3D classification and refinement of ΔMtr4 90S particles.
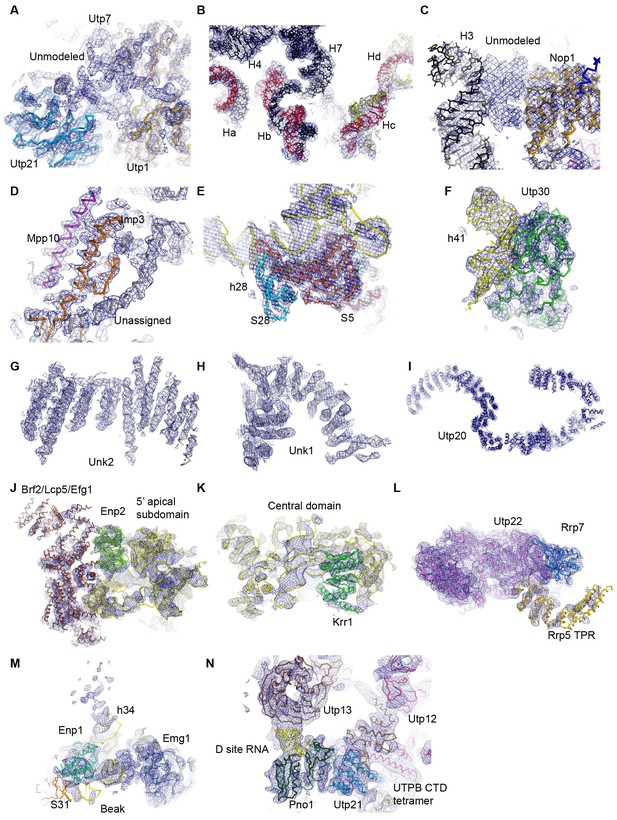
Representative densities fitted with structural models.
(A) Unmodeled densities near Utp21 and Utp1. (B) The 5' ETS and U3 RNA. (C) An unmodeled density between H3 and Nop1. (D) Imp3 and Mpp10 located at the interior of the structure. (E) The 3' basal subdomain. (F) Utp30 and helix 41. (G) The unassigned protein Unk2. (H) The unassigned protein Unk1. (I) Utp20. (J) The 5' apical subdomain, Enp2 and bulky unassigned densities adjacent to Enp2. (K) The central domain and Krr1. (L) Utp22, Rrp7 and the TRP domain of Rrp5. (M) The 3' beak subdomain. (N) The D site RNA, Pno1 and the UTPB complex. The ΔMtr4 cryo-EM density map is displayed in A-G and the ΔDhr1 cryo-EM density map (state 1) is displayed in H–N.
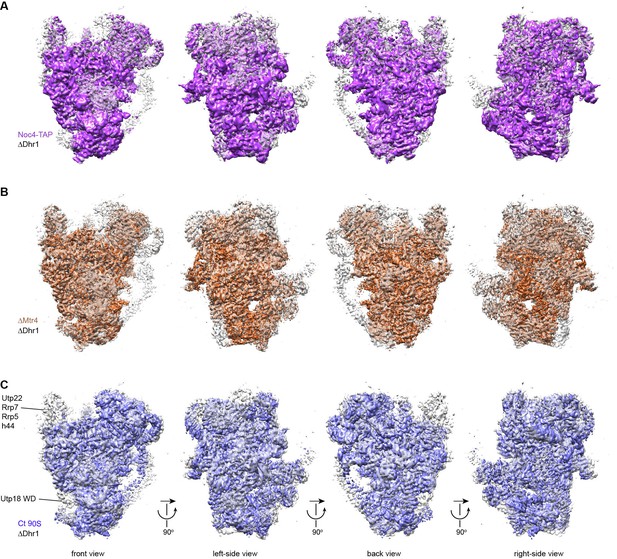
Alignment of cryo-EM maps of 90S.
(A) Alignment of the ΔDhr1 (grey) and Noc4-TAP (purple) 90S maps. Four orthogonal views are displayed. (B) Alignment of the ΔDhr1 (grey) and ΔMtr4 90S (orange) maps. (C) Alignment of the ΔDhr1 (grey) and Ct 90S (blue) maps. The densities missing in the Ct 90S map are labeled.
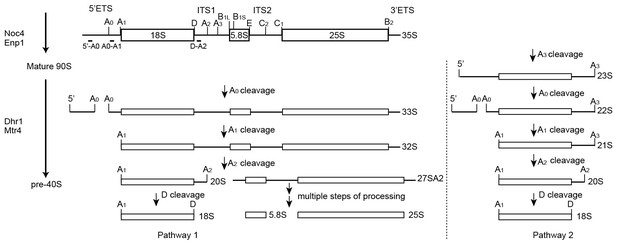
Processing pathway of 18S rRNA.
The 35S pre-rRNA is cleaved sequentially at the A0, A1 and A2 sites to generate the 20S and 27SA2 pre-rRNAs. Alternatively, initial cleavage at site A3 yields a 23S pre-rRNA, which is then processed at the A0, A1 and A2 sites. The 20S pre-rRNA maturates into 18S rRNA after D site cleavage. Noc4 and Enp1 associate with 90S during its co-transcriptional assembly. Dhr1 and Mtr4 likely associate after the A0 site is cleaved.
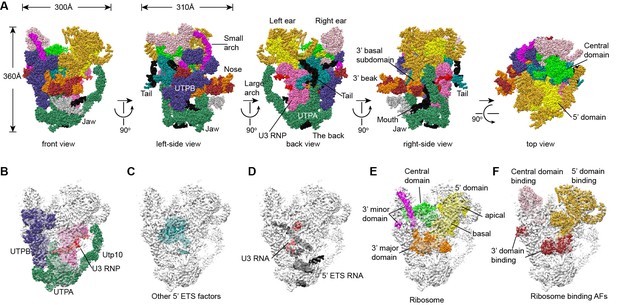
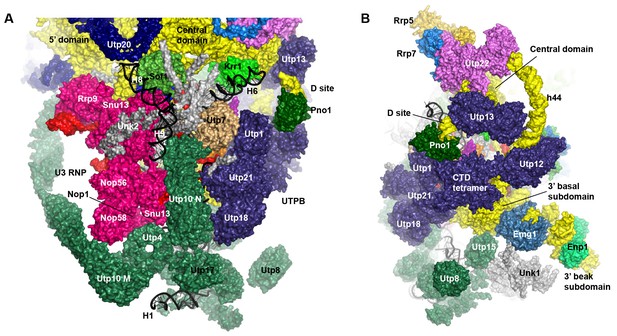
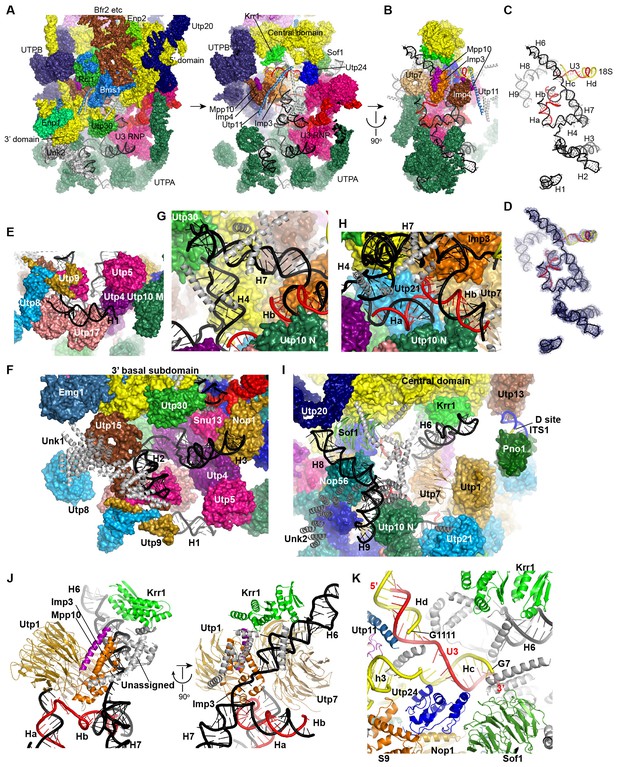
Structural organization of 90S.
(A) Space-filling model of 90S in five perpendicular views. Substructures are color-coded as described below and other unassigned proteins are grey. Major structural features are indicated. (B) UTPA (sea green), UTPB (dark slate blue) and U3 snoRNP (hot pink). (C) Other 5' ETS proteins including the unassigned proteins around 5' ETS RNA (dark cyan). (D) 5' ETS (black) and U3 (red) RNA. (E) The 5' domain (yellow), central domain (green), 3' major domain (orange) and 3' minor domain (magenta) of small subunit ribosomes. (F) The AFs bound to the 5' domain (goldenrod), central domain (pink), 3' major domain (brown) and 3' minor domain (brown). The substructures in B–F are shown in front view and with the ΔDhr1 density map.
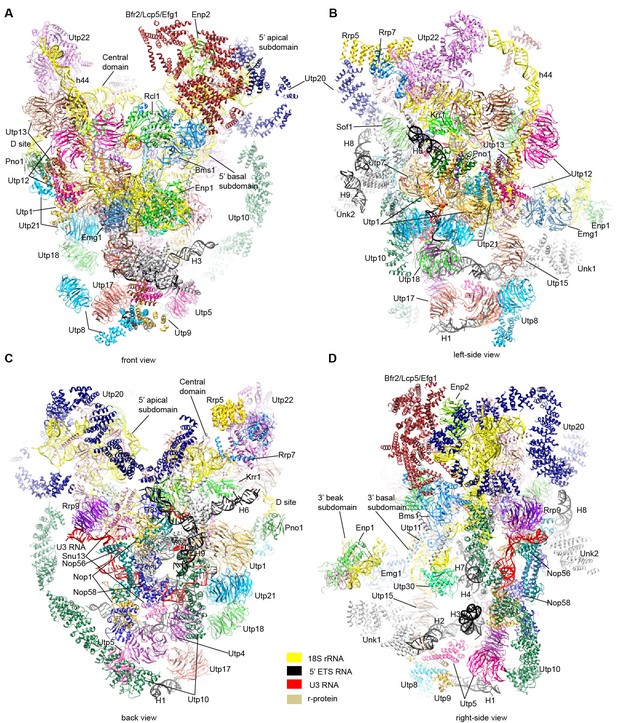
Structural model of 90S.
(A–D) Structural model of 90S in the front (A), left-side (B), back (C) and right-side (D) view. The 18S, 5' ETS and U3 RNAs are colored yellow, black and red, respectively. The assigned AFs are color coded, all r-proteins are wheat and unassigned proteins are grey.
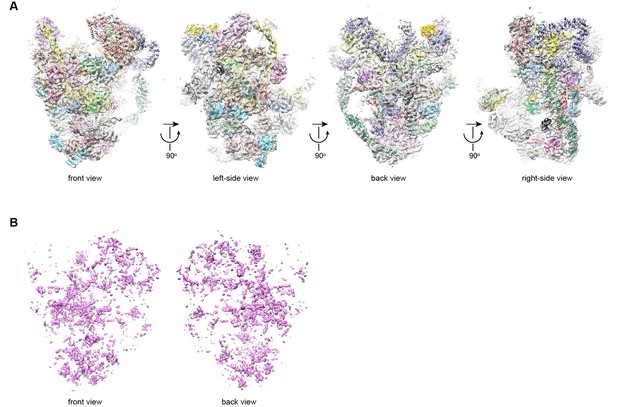
Structural model of 90S.
(A) Fitting of the 90S model in the ΔDhr1 cryo-EM density map. Four orthogonal views are shown. (B) The unmodeled densities in the ΔDhr1 map.
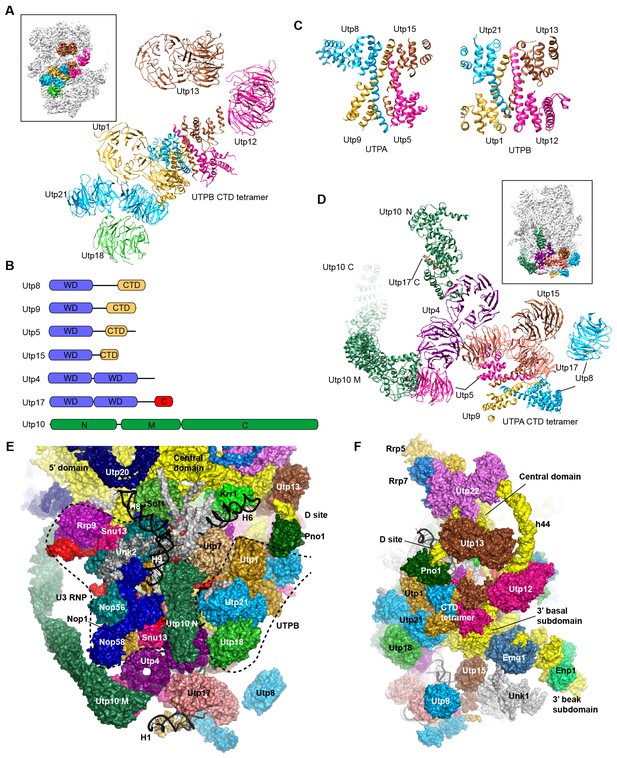
Structure of UTPB and UTPA.
(A) Structure of UTPB in 90S. The insert shows the displayed region in the ΔDhr1 map. (B) Domain diagram of UTPA proteins. (C) UTPB and UTPA share a homologous CTD tetramer. (D) Structure of UTPA in 90S. The insert shows the displayed region in the ΔDhr1 map. (E) UTPA, UTPB, U3 snoRNP and additional 5' ETS factors form the back. UTPB and U3 snoRNP are marked with dotted lines. (F) Interface of UTPA and UTPB to the nascent ribosome.
Structure of UTPB and UTPA.
(A–B) Duplication of Figure 4E and F, but all the proteins of UTPA, UTPB and U3 RNP are colored in sea green, deep sky blue and hot pink, respectively.
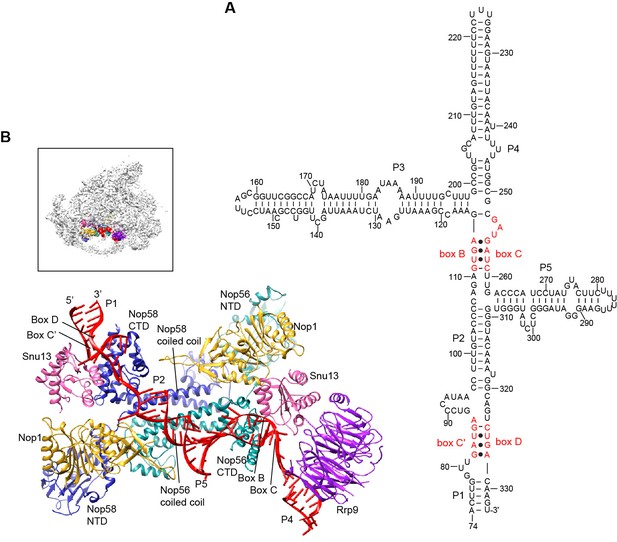
Structure of U3 snoRNP.
(A) Secondary structure model of the 3' domain of U3 RNA. Paired regions are labeled as P1 to P5. The box C', B, C and D motifs are red. (B) Structure of U3 snoRNP in 90S. The insert shows the displayed region in the ΔDhr1 map.
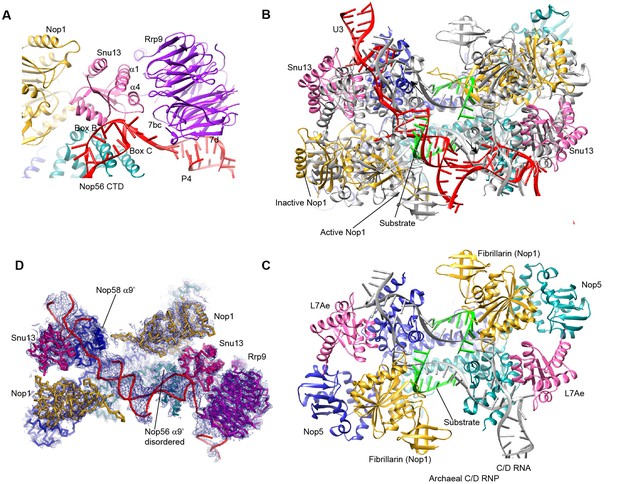
Structure of U3 snoRNP.
(A) Interaction of Rrp9 with Snu13 and U3 RNA. (B) The P2 helix structurally mimics a guide-substrate duplex. The U3 snoRNP structure is aligned with the substrate-bound structure of archaeal C/D RNP in the active state by the coiled-coil domain of Nop5. U3 snoRNP is color-coded. The archaeal C/D RNP structure is colored in grey except that the bound substrates are colored in green. (C) Structure of C/D RNP (PDB code 3PLA). Each component is color-coded. (D) Fitting of U3 snoRNP structure in the ΔMtr4 density map.
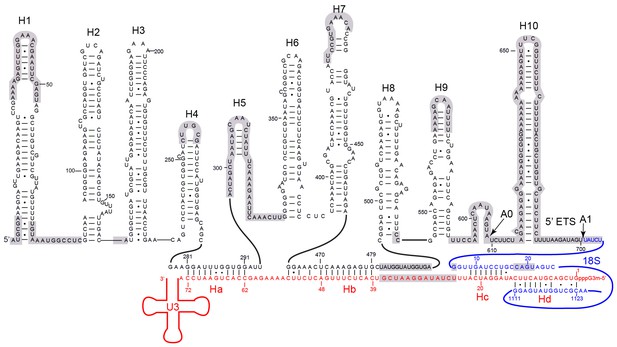
Secondary structure model of the 5' ETS and the 5' domain of U3 RNA.
Paired regions of the 5' ETS RNA are labeled as H1 to H10. The 5' ETS and 18S RNA are numbered independently. Four hybrid duplexes of U3 observed in the 90S structure are named Ha, Hb, Hc and Hd. The grey-shaded sequences are not modeled.
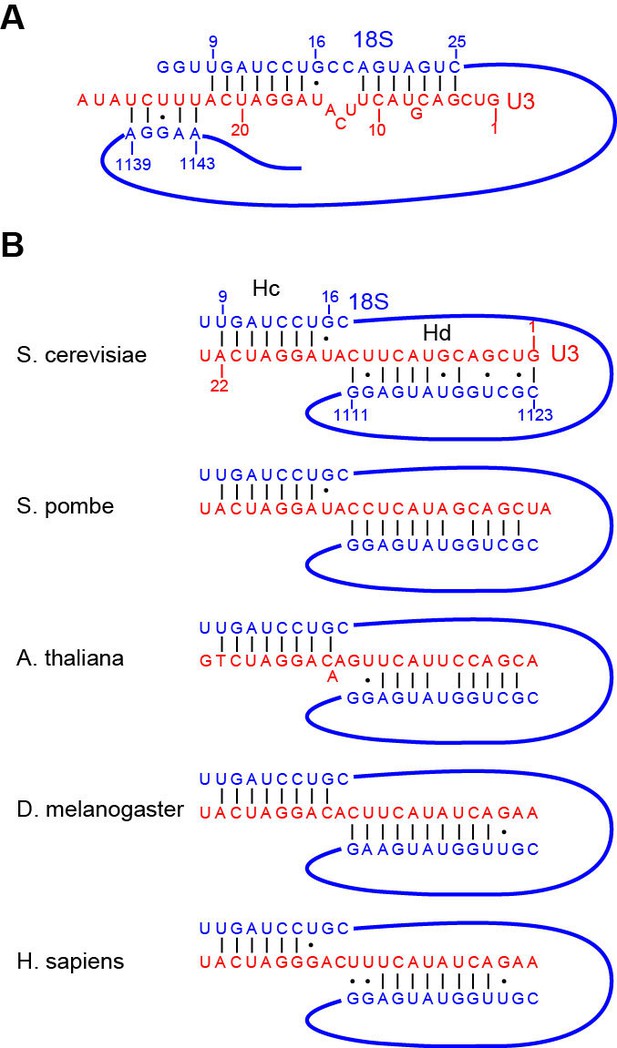
Interaction of U3 with 18S RNA.
(A) The previous model of U3-18S interaction. The U3 RNA is red and the 18S RNA is blue. (B) The new model of U3-18S interaction. The base pairing interaction between U3 and the helix 27 sequence of 18S is conserved in S. cerevisiae, S. pombe, A. thaliana, D. melanogaster and H. sapiens.
Structure and interaction of 5' ETS and the 5' domain of U3 RNA.
(A) Open-box view of 5' ETS and U3 RNA. The left view shows an intact 90S structure. The right view is resulted from removal of the 5' and 3' domain of small subunit, Utp30, Enp1, Unk2, Utp20, Bms1, Rcl1, Enp2 and Bfr2. The unassigned structures are colored grey. (B) A 90° rotation of A. UTPB is additionally removed. (C) Structure of 5' ETS and the 5' domain of U3 RNA. Same orientation as in B. (D) The ΔMtr4 EM density for 5' ETS and the 5' domain of U3 RNA. (E) H1. (F) H2 and H3. A view of the mouth. (G) H4 and H7. (H) Ha and Hb. (I) H6, H8 and H9. (J) Imp3 binds at the junction of H6, H7, Ha and Hb. Two perpendicular views are shown. (K) Hc and Hd. The 5' nucleotides G7 and G1111 of the 18S strands in Hc and Hd are labeled.
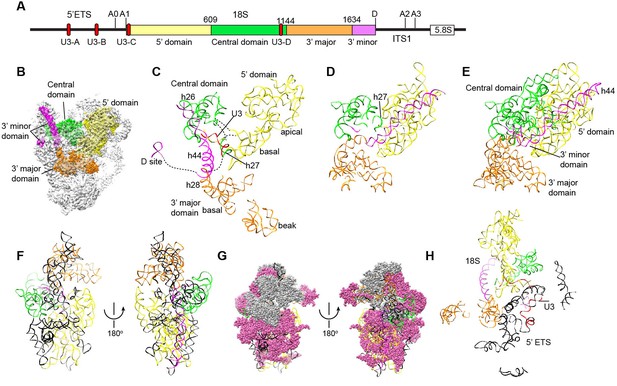
Nascent ribosome in 90S.
(A) Diagram of pre-rRNA. 25S RNA is omitted. Processing sites, four 18S domains and four U3 binding sites are labeled. (B) Assembled ribosome in 90S. The 5' domain, central domain, 3' major domain and 3' minor domain of 18S and their associated r-proteins are colored in yellow, green, orange and magenta, respectively. The color theme is also used in other panels. (C) Structure of 18S RNA in 90S. Dotted lines show connections between major domains. (D) The 18S RNA detected in 90S is mapped to 40S structure. The central domain is aligned to the structure in both panels B and C. (E) 18S RNA in 40S structure. Same view as in D. (F–G) The 18S RNA (F) and r-proteins (G) observed in 90S are mapped to 40S structure. Undetected RNAs and proteins are colored black and grey. Detected r-proteins are colored deep pink. (H) Structural separation between the 18S and 5' ETS RNA.
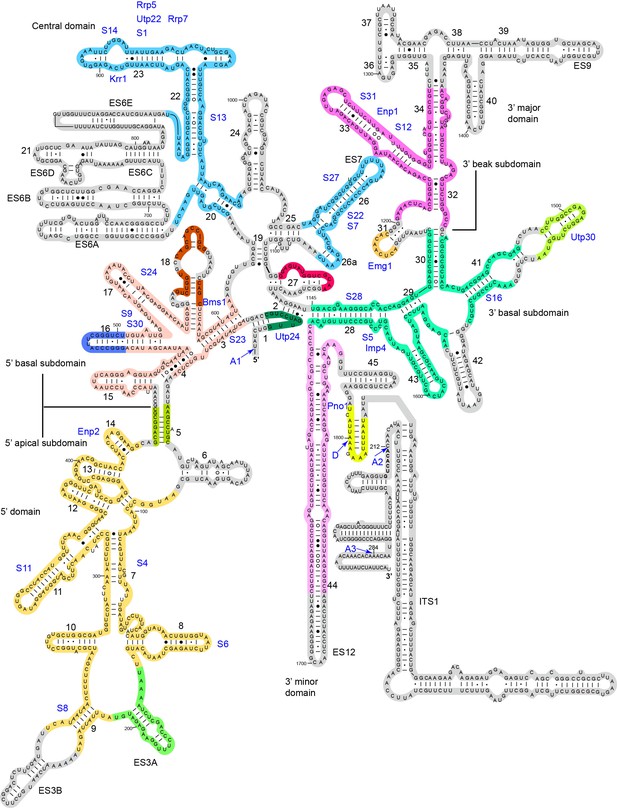
Assembly of 18S RNA in 90S.
The secondary structure model is shown for 18S RNA in the mature ribosome and part of the ITS1. The modeled regions are shaded with different colors and the unmodeled regions are shaded grey. Except for the two U3 binding sites on the 5' end of 18S and helix 27, the RNA sequences of the same color generally assemble into native-like local structures in 90S. The differently colored RNA segments do not maintain a native alignment relative to each other. The pre-rRNA processing sites and helix numbers are labeled. The r-proteins and closely associated AFs are placed near their major binding site.
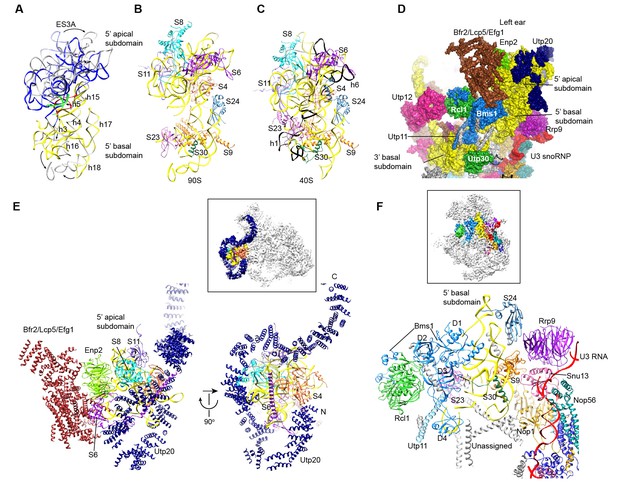
Structure of the 5' domain in 90S.
(A) The 5' domain RNA in 90S (grey and green) and 40S (yellow, red and green) are aligned by the basal subdomain. Shifted RNA elements are marked with arrows. (B–C) The assembled 5' domain structure in the 90S (B) and 40S (C) ribosome. RNAs disordered in 90S are colored black in C. Same orientation as in A. (D) Structure of the 5' domain and associated AFs shown in surface view. The beak region is removed for clarity. (E) Interactions of the 5' apical subdomain with AFs are shown in two perpendicular views. The insert shows the displayed region in the ΔDhr1 map. (F) Interactions of the 5' basal subdomain with AFs. The insert shows the displayed region in the ΔDhr1 map.
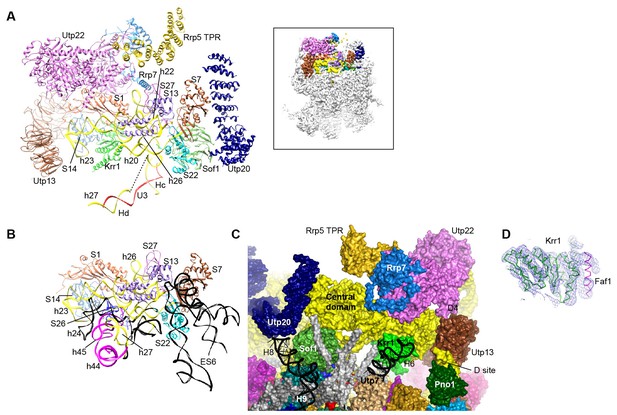
Structure of the central domain in 90S.
(A) Structure of the central domain with bound AFs in 90S. The insert shows the displayed region in the ΔDhr1 map. (B) The central domain structure in the mature 40S is shown in the same view as A. The 18S RNA regions not detected in 90S are colored black. (C) Interface of the central domain with AFs in surface view. The orientation is nearly opposite to A. (D) Structure of the Krr1-Faf1 complex is fitted into the ΔDhr1 density map.
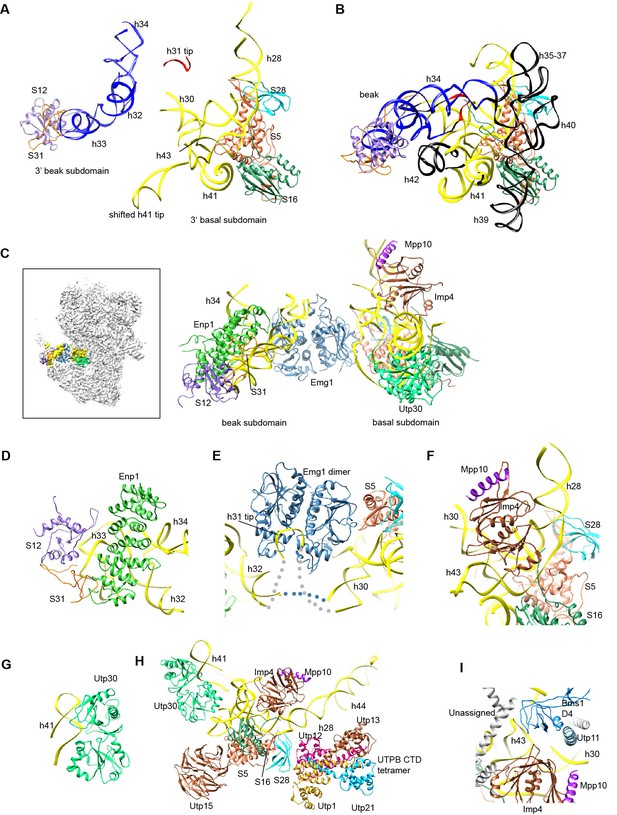
Structure of the 3' major domain in 90S.
(A) The 3' major domain structure in 90S. RNA segments are color-coded. (B) The 3' major domain structure in 40S. Aligned to the 3' basal subdomain in A. The RNA regions undetected in 90S are colored black and other regions are colored as their counterparts in A. Only the 5 r-proteins detected in 90S are displayed. (C) The 3' major domain with closely associated AFs. The insert shows the displayed region in the ΔDhr1 map. (D) Interaction of Enp1 with the beak. (E) Interaction of Emg1 with the 3' major domain. (F) Interaction of Imp4 with the 3' basal subdomain. (G) Interaction of Utp30 with helix 41. (H) Interaction of the 3' basal subdomain with UTPA and UTPB components. (I) Interaction of the 3' basal subdomain with Bms1 and Utp11.
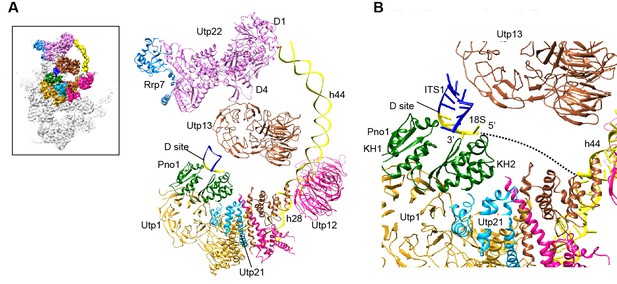
Structure of the 3' minor domain in 90S.
(A) Assembly of helix 44 and the D site hairpin. The 18S and ITS1 RNA are colored yellow and blue. The insert shows the displayed region in the ΔDhr1 map. (B) Structure of Pno1 and the D site hairpin. The dotted line indicates a potential path of helix 45.
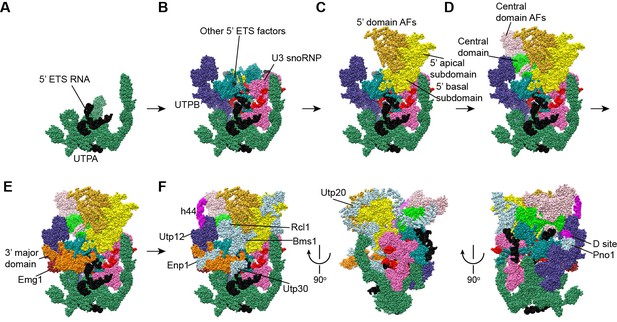
Stepwise assembly of 90S.
(A–F) Components are added by their known assembly order. Assembly of UTPA (A), UTPB, U3 snoRNP and other 5' ETS factors (B), the 5' domain and its AFs (C), the central domain and its AFs (D), the 3' major domain and its AFs (E) and the 3' minor domain and the late factors (F). The view is rotated by −45° along y axis from the front view. The fully assembled 90S is additionally shown in two perpendicular views. Late AFs are labeled in F.
Videos
Structure of 90S.
The first two sections illustrate the ΔDhr1 (state 1) map and the structural model of 90S and the last section shows only the structure of nascent ribosome, 5' ETS and U3 RNA.
Additional files
-
Supplementary file 1
Components and modeling of yeast 90S structure.
- https://doi.org/10.7554/eLife.22086.028
-
Supplementary file 2
Chemical crosslinking and mass spectrometry data for the ΔMtr4/Enp1-TAP sample.
Sheet one stores the crosslinked peptides. Sheet two shows the annotated intermolecular crosslinks. Sheet three shows the intramolecular crosslinks. The structural consistency for crosslinks between two assigned proteins is judged by the distance (<30 Å) between crosslinked sites. If a crosslinked site is unmodeled or undetermined in structure, the position of neighboring residues or domains is considered.
- https://doi.org/10.7554/eLife.22086.029
-
Supplementary file 3
Statistics of data collection, structural refinement and model validation.
- https://doi.org/10.7554/eLife.22086.030





