Srs2 promotes synthesis-dependent strand annealing by disrupting DNA polymerase δ-extending D-loops
Figures
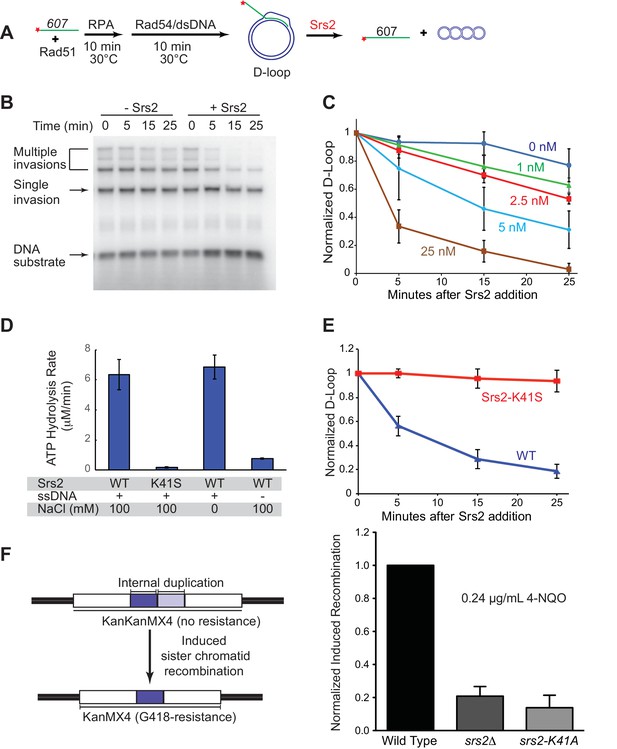
Srs2 disrupts D-loops in a concentration-, time-, and helicase activity-dependent manner.
(A) D-loop disruption assay. Fully homologous ssDNA with 607 nucleotides was used as substrates in B and C. Rad51 (0.2 µM) was incubated with 1 nM ssDNA substrate (Rad51/nt = 1:3) for 10 min at 30°C. RPA (33 nM) was added for another 10 min incubation before the addition of 84 nM Rad54 and 7 nM supercoiled plasmid dsDNA (21 µM bp). (B) Time course of Srs2 titration into pre-formed D-loops produced by Rad51 and Rad54. Srs2 (0 or 5 nM) was added and incubated for various time at 30°C. Samples were taken at 0, 5, 15, and 25 min and quenched by SDS/Proteinase K treatment. (C) Quantitation of normalized total D-loop yield in (B) and additional titrations. Absolute initial D-loop yields were between 50% and 75%. (D) Srs2-K41S is deficient in ATP hydrolysis. Rates of ATP hydrolysis were determined for 5 nM wild type Srs2 (WT) or catalytic-deficient Srs2-K41S (K41S) in D-loop reaction buffer containing either 0 or 100 mM NaCl, with or without 10 µM φX174 ssDNA as cofactor. (E) Time course of D-loop disruption by 5 nM of either wild type Srs2 (WT) or catalytic-deficient Srs2-K41S (K41S). Plotted are means ± standard deviation from n = 3. Absolute initial D-loop yields were between 44% and 72%. (F) Induced sister chromatid recombination assay. The normalized (wild type = 100%) induced recombination frequencies are shown at 0.24 µg/mL 4-NQO as means ± standard deviations from three experiments for wild type, srs2△, and srs2-K41A (see Table 1). The frequency of spontaneous G418R cells was subtracted from the induced recombination frequencies. The full 4-NQO dose response is shown in Figure 1—figure supplement 2.
-
Figure 1—source data 1
Source data for Figure 1C.
- https://doi.org/10.7554/eLife.22195.003
-
Figure 1—source data 2
Source data for Figure 1D.
- https://doi.org/10.7554/eLife.22195.004
-
Figure 1—source data 1
Source data for Figure 1E.
- https://doi.org/10.7554/eLife.22195.005
-
Figure 1—source data 4
Source data for Figure 1F and Figure 1—figure supplement 2A–D.
- https://doi.org/10.7554/eLife.22195.006
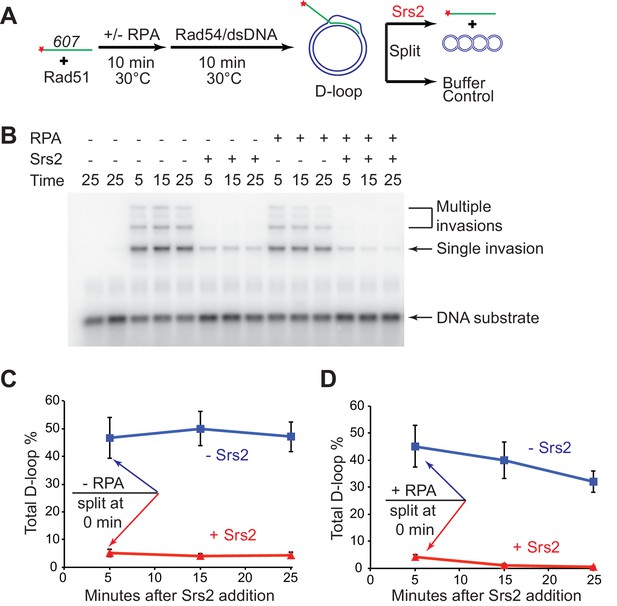
D-loop disruption by Srs2 does not depend on RPA.
(A) D-loop disruption assay based on ssDNA substrate mimicking physiological length. Fully homologous ssDNA with 607 nucleotides was used as the substrate to produce D-loops. 0.2 µM Rad51 was incubated with 1 nM ssDNA substrates (Rad51/nt = 1:3) for 10 min at 30°C. 33 nM RPA was added for another 10 min incubation before the addition of 84 nM Rad54 (84 nM) and 7 nM supercoiled plasmid dsDNA (21 µM bp). Reactions with or without RPA were individually split into buffer containing Srs2 or buffer. (B) Time course of Srs2 disruption of pre-formed D-loops produced by Rad51 and Rad54 in the presence and absence of RPA. The final concentration of Srs2 is 25 nM. Samples were taken at 0, 5, 15, and 25 min and quenched by SDS/Proteinase K treatment. (C) Quantitation of total D-loops in the absence of RPA with or without Srs2. (D) Quantitation of total D-loops in the presence of RPA with or without Srs2. Plotted are means ± standard deviation from n = 3.
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1.
- https://doi.org/10.7554/eLife.22195.008
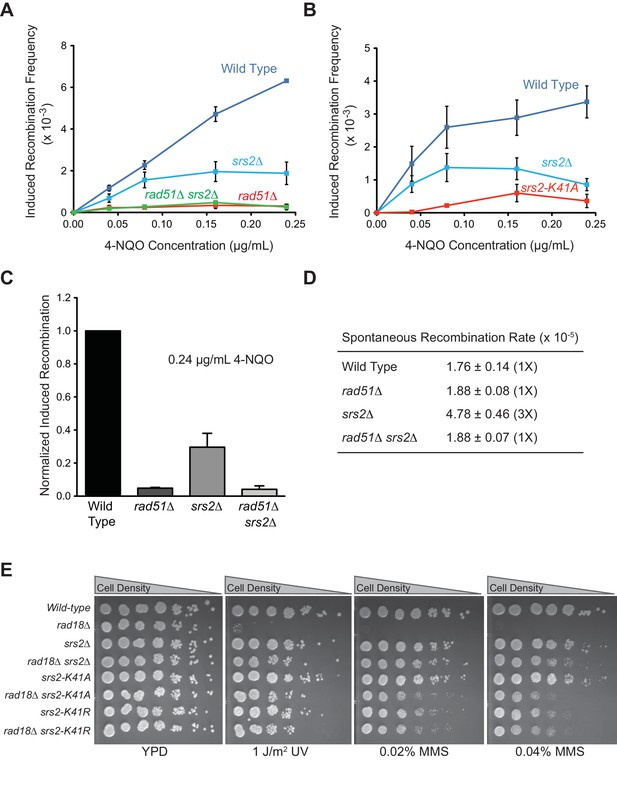
ATPase activity of Srs2 is required for both pro- and anti-recombination function and required for 4-NQO-induced recombinational repair, and responsible for the extreme UV and MMS sensitivities of rad18△ strain.
(A) Induced recombination frequency for wild type (WDHY5473), srs2△ (WDHY5475), rad51△ (WDHY5474), and rad51△ srs2△ (WDHY5476). (B) Induced recombination frequency of wild type, srs2△, and srs2-K41A. Induced recombination frequencies of wild type and mutant cells in dose response to 4-NQO (0.04, 0.08, 0.16, 0.24 µg/mL) are shown as means ± standard deviations from three experiments. The frequency of spontaneous G418R cells was subtracted from the induced recombination frequencies. (C). Induced sister chromatid recombination assay. Bar graph representation of normalized (wild type = 100%) induced recombination frequency at 0.24 µg/mL 4-NQO for wild type (WDHY5473), srs2△ (WDHY5475), rad51△ (WDHY5474), and rad51△ srs2△ (WDHY5476). (D). Spontaneous recombination rates of strains in (C). The fold increase is shown in brackets. (E) UV and MMS sensitivities of wild type (WDHY3429), rad18Δ (WDHY3514), srs2Δ (WDHY3858), rad18Δ srs2Δ (WDHY4083), srs2-K41A (WDHY3851), rad18Δ srs2-K41A (WDHY4082), srs2-K41R (WDHY4130), and rad18△ srs2-K41R (WDHY4141) at 30°C in the serial dilution assays.
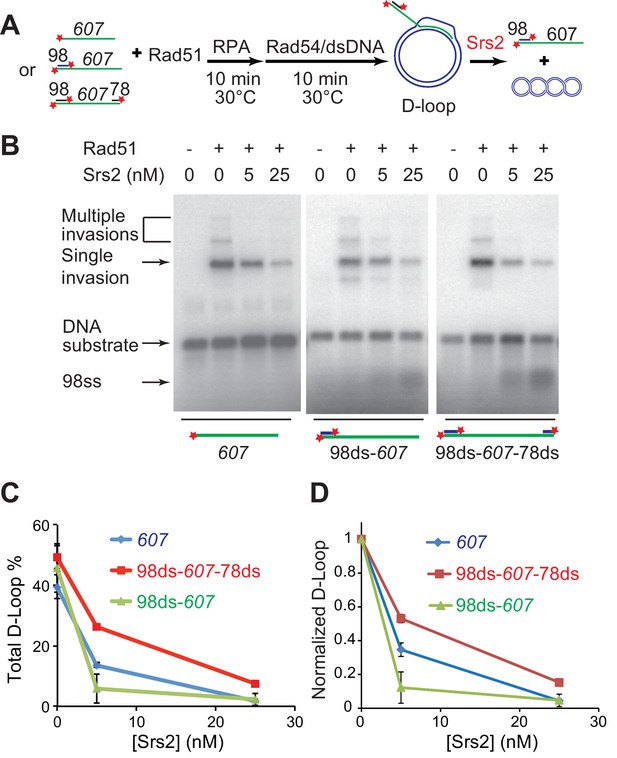
Srs2 disrupts various D-loops produced from ssDNA substrates mimicking physiological length and structure.
(A) D-loop disruption assay and substrates. Rad51 (0.2 µM) was incubated with 1 nM ssDNA substrates (Rad51/nt = 1:3) for 10 min at 30°C. 33 nM RPA was added for another 10 min incubation before the addition of 84 nM Rad54 (84 nM) and 7 nM supercoiled plasmid dsDNA (21 µM bp). Srs2 (0, 5, or 25 nM) was added and incubated for 10 min at 30°C before the reaction was stopped by SDS/Proteinase K. (B) Srs2 titration in D-loop disruption assays with three different DNA constructs, 3' tailed DNA with 5' heterology, ssDNA with full homology, gapped DNA with both 5' and 3' heterologies. (C) Quantitation of total D-loops from (B). (D) Quantitation of normalized D-loops from (B) setting initial D-loop yield as 100%. Plotted are means ± standard deviation from n = 3.
-
Figure 2—source data 1
Source data for Figure 2.
- https://doi.org/10.7554/eLife.22195.011
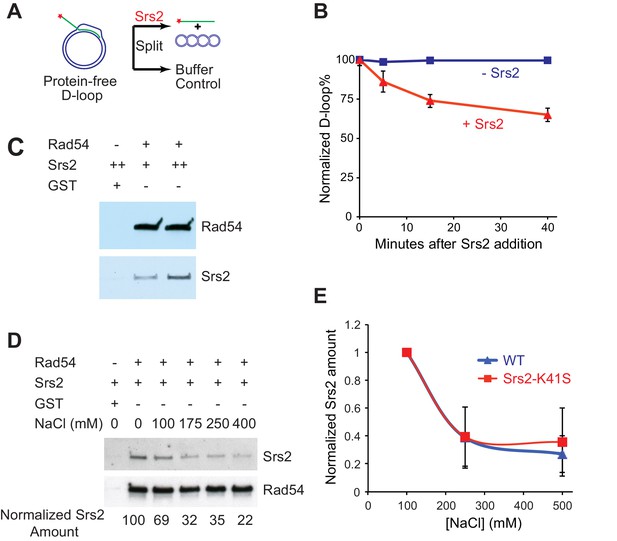
Srs2 disrupts Rad51/Rad54 reconstituted D-loops with higher efficiency than protein-free D-loops.
(A) D-loop disruption assay using purified protein-free D-loops generated by Rad51 and Rad54. (B) Protein-free D-loops were split into buffer containing Srs2 or no protein and incubated at 30°C. Samples were taken at 0, 5, 15, and 40 min, and the final concentration of Srs2 is 15 nM. (C) GST-Rad54 physically interacts with Srs2. 17.5 nM GST-Rad54 or 175 nM GST were incubated with either 8.75 nM or 17.5 nM of Srs2 for 1 hr before pulldown. (D) The protein interaction between Rad54 and Srs2 is sensitive to increasing ionic strength. GST-Rad54 and Srs2 were formed in buffer containing 0, 100, 175, 250, or 400 mM NaCl before pulldown. (E) Srs2-K41S interacts with Rad54 with similar salt sensitivity, compared to wild type Srs2. Both wild type Srs2 and Srs2-K41S were allowed to form complex with GST-Rad54 in buffer containing 0, 250, and 500 mM NaCl before pulldown. Plotted are means ± standard deviation from n = 3. In (D) and (E), 31.3 nM GST-Rad54 or 313 nM GST (GE Healthcare) were incubated with 31.3 nM of Srs2 or Srs2-K41S in the same buffer containing indicated amount of NaCl for 1 hr before pulldown. Pulldown Srs2 amount in buffer containing 0 mM NaCl was normalized to 100 (D) or 1 (E).
-
Figure 3—source data 1
Source data for Figure 3B.
- https://doi.org/10.7554/eLife.22195.013
-
Figure 3—source data 2
Source data for Figure 3E.
- https://doi.org/10.7554/eLife.22195.014
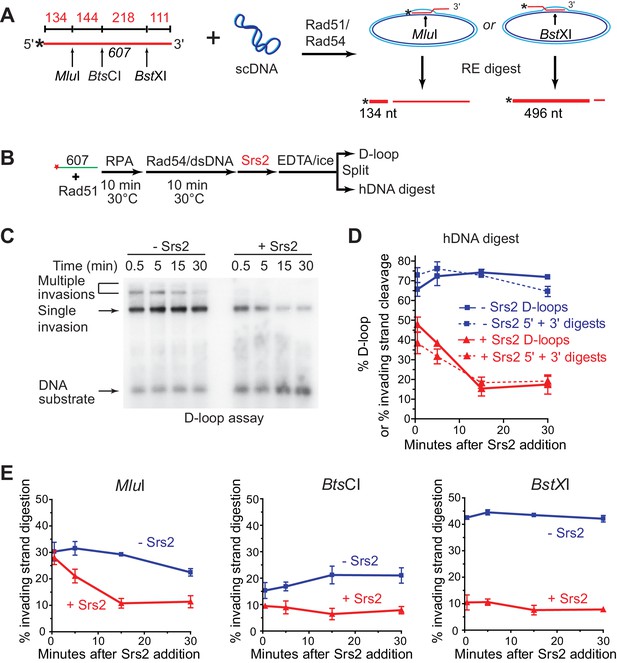
Srs2 preferentially disrupts D-loops with 3' proximal hDNA.
(A) hDNA detection assay. Note that since the BstXI and MluI sites are 362 nt apart, they cannot be incorporated into the same plasmid DNA molecule in a contiguous hDNA tract, and therefore the two sites are mutually exclusive and report on the formation of D-loop products with 3' or 5' proximal hDNA content. (B) Scheme of the experiment. (C) Representative D-loop assay gel. Times points are post-Srs2 (5 nM) or dilution buffer control addition, starting 10 min after the initiation of the D-loop reaction with Rad54 protein and homologous donor plasmid addition. (D) Quantitation of D-loops and the sum of the 3' and 5' proximal digestion products. (E) Quantitation of hDNA digestion at three sites on the invading DNA molecule, as indicated in the schematic at top. Note that the middle BtsCI site is close enough to the two other sites that it is not mutually exclusive for incorporation into a fraction of the 5' or 3' proximal hDNA products. Plotted are means ± standard deviation from n = 3.
-
Figure 4—source data 1
Source data for Figure 4.
- https://doi.org/10.7554/eLife.22195.016
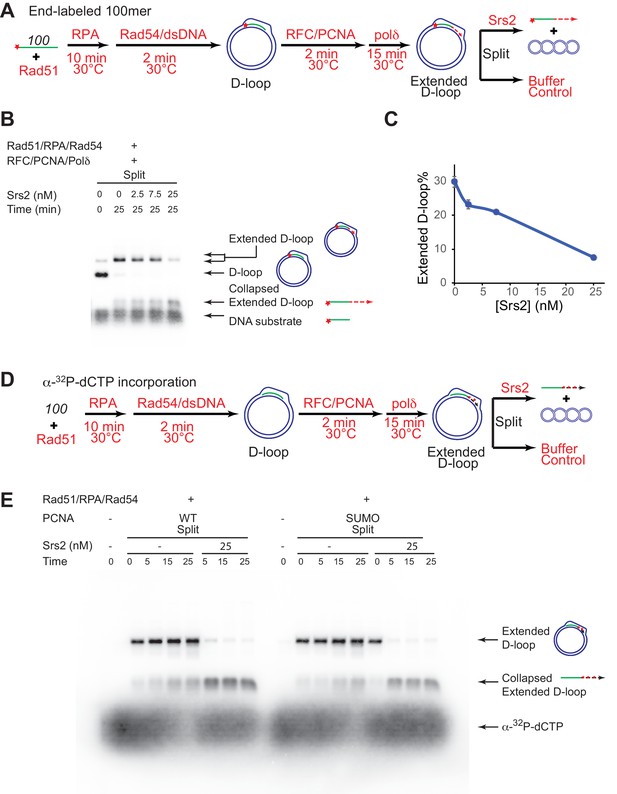
Srs2 disrupts extended D-loops produced by DNA polymerase δ, in a reconstituted assay system containing RFC, PCNA, Rad51 and Rad54.
(A) Extended D-loop disruption assay. Fully homologous 100mer ssDNA was used as substrates. 0.2 µM Rad51 was incubated with 6 nM ssDNA substrates (Rad51/nt = 1:3) for 10 min at 30°C. RPA (165 nM) was added for another 10 min incubation before the addition of 84 nM Rad54 and 7 nM supercoiled plasmid dsDNA (21 µM bp). After 2 min, 20 nM RFC and 20 nM PCNA were added and incubated for another two min at 30°C before the addition of 7 nM DNA polymerase δ. After 15 min, different amount of Srs2 or storage buffer were added and samples at 25 min were taken and stopped by SDS/Proteinase K treatment. (B) Titration of Srs2 (0, 2.5, 7.5, or 25 nM final concentration) in extended D-loop disruption. (C) Quantitation of extended D-loops in B). Plotted are means ± standard deviation from n = 3. (D) Extended D-loop disruption assay using α-32P-dCTP incorporation. Experimental setup and protein concentrations are identical as in A), except that α-32P-dCTP was included in the buffer and 100mer ssDNA was not radiolabeled. (E) Time course of extended D-loop disruption by Srs2, in the presence of PCNA and SUMO-PCNA. After 15 min, Srs2 (25 nM final concentration) or storage buffer were added and samples at 0, 5, 15, and 25 min were taken and stopped by SDS/Proteinase K treatment.
-
Figure 5—source data 1
Source data for Figure 5C.
- https://doi.org/10.7554/eLife.22195.018
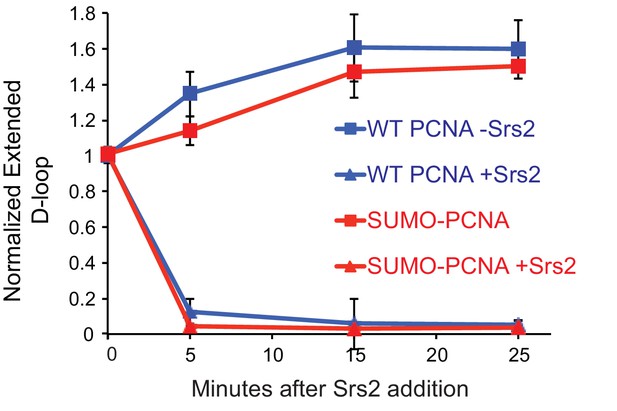
Srs2 disrupts WT PCNA and SUMO-PCNA extending D-loops with similar kinetics at optimal conditions.
Quantitation of normalized extended D-loops in Figure 5E. Note that the yield of extended D-loops is identical with either WT PCNA or SUMO-PCNA. The intensities of the extended D-loops were normalized against the signal of the extended D-loop produced with wild type PCNA without Srs2 at 0 min in each repeat. Plotted are means ± standard deviation from n = 3.
-
Figure 5—figure supplement 1—source data 1
Source data for Figure 5—figure supplement 1.
- https://doi.org/10.7554/eLife.22195.020
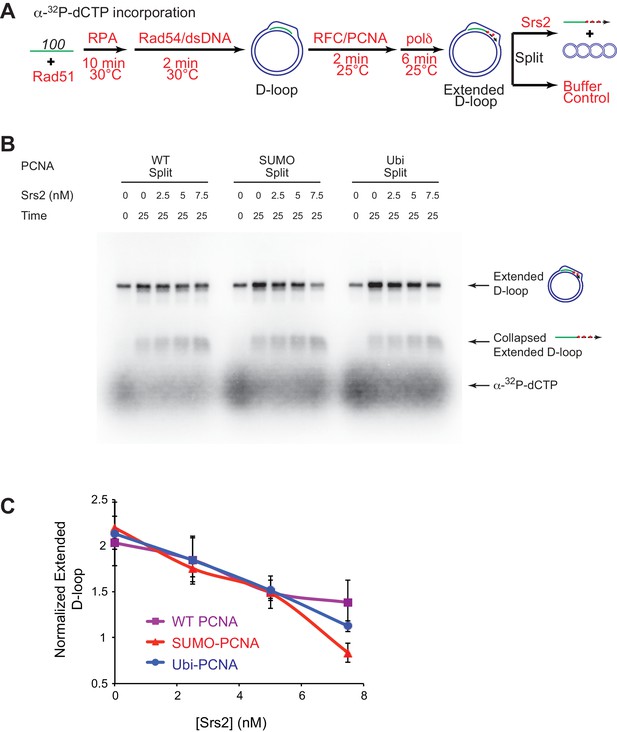
Slight preference of Srs2-mediated disruption for PCNA-SUMO containing reconstituted reactions compared to unmodified or ubiquitinated PCNA.
(A) Extended D-loop disruption assay using α-32P-dCTP incorporation. Fully unlabeled and homologous 100mer ssDNA was used as substrates, and α-32P-dCTP was included in the buffer. 0.2 µM Rad51 was incubated with 6 nM ssDNA substrates (Rad51/nt = 1:3) for 10 min at 30°C. RPA (165 nM) was added for another 10 min incubation before the addition of 84 nM Rad54 and 7 nM supercoiled plasmid dsDNA (21 µM bp). After 2 min, 20 nM RFC and 20 nM PCNA, PCNA-SUMO, or PCNA-Ubi, were added and temperature was lowered to 25°C. DNA polymerase δ (7 nM) was added after two more min and incubated for 6 min at 25°C. Reactions with PCNA, SUMO-PCNA, and Ubi-PCNA were individually split into Srs2 or buffer only control. Final concentrations of Srs2 are 0, 2.5, 7.5, or 25 nM. Samples at 25 min were taken and stopped by SDS/Proteinase K treatment. (B) Titration of Srs2 in extended D-loop disruption. (C) Quantitation of normalized extended D-loops in (B). The intensities of the extended D-loops were normalized against the signal of the extended D-loop produced with wild type PCNA without Srs2 at 0 min in each repeat. Plotted are means ± standard deviation from n = 3.
-
Figure 6—source data 1
Source data for Figure 6C.
- https://doi.org/10.7554/eLife.22195.022
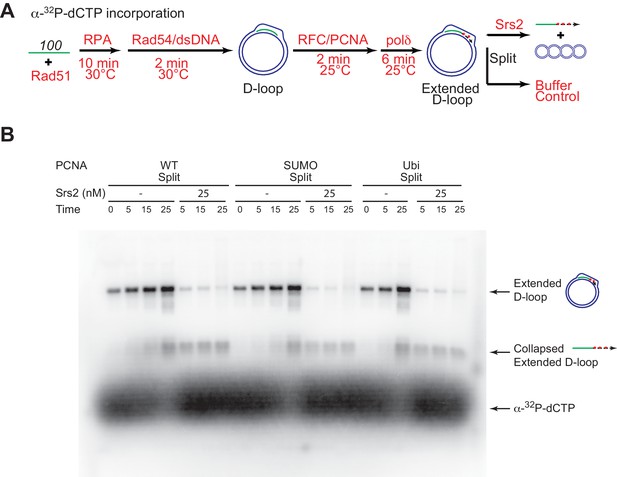
High concentrations of Srs2 are equally proficient to disrupt D-loops extending with different forms of PCNA.
(A) Extended D-loop disruption assay using α-32P-dCTP incorporation. Fully unlabeled and homologous 100mer ssDNA was used as substrates, and α-32P-dCTP was included in the buffer. 0.2 µM Rad51 was incubated with 6 nM ssDNA substrates (Rad51/nt = 1:3) for 10 min at 30°C. RPA (165 nM) was added for another 10 min incubation before the addition of 84 nM Rad54 and 7 nM supercoiled plasmid dsDNA (21 µM bp). After 2 min, 20 nM RFC and 20 nM PCNA, SUMO-PCNA, or Ubi-PCNA, were added and temperature was lowered to 25°C. DNA polymerase δ (7 nM) was added after two more min and incubated for 6 min at 25°C. Reactions with PCNA, SUMO-PCNA, and Ubi-PCNA were individually split into Srs2 or buffer only control. Samples at 0, 5, 15, and 25 min were taken and stopped by SDS/Proteinase K treatment. (B) Time course of extended D-loop disruption by Srs2.
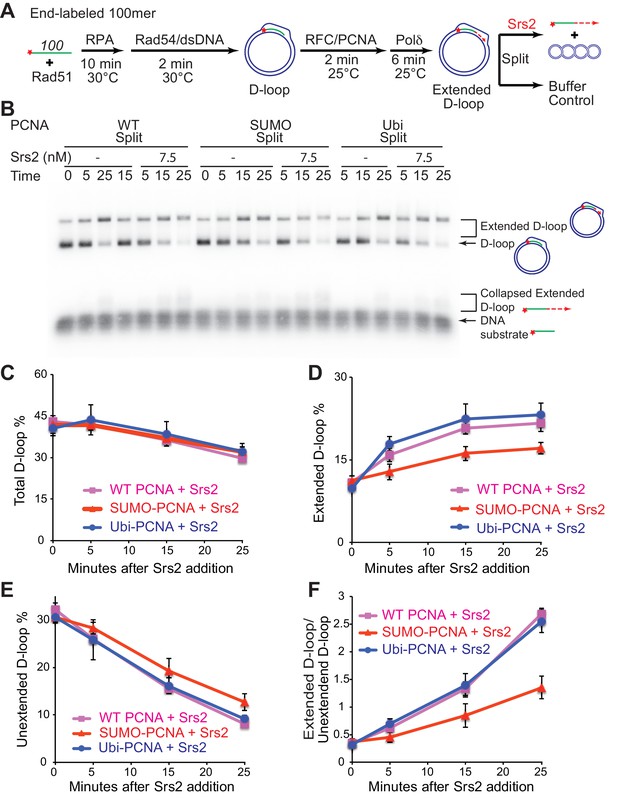
Srs2 prefers to disrupt extended D-loop over unextended D-loop when SUMO-PCNA is present in a reconstituted system compared to unmodified or Ubi PCNA.
(A) Extended D-loop disruption assay. Fully homologous 100mer ssDNA was used as substrates. Rad51 (0.2 µM) was incubated with 6 nM ssDNA substrates (Rad51/nt = 1:3) for 10 min at 30°C. RPA (165 nM) was added for another 10 min incubation before the addition of 84 nM Rad54 and 7 nM supercoiled plasmid dsDNA (21 µM bp). After 2 min, 20 nM RFC and 20 nM PCNA, SUMO-PCNA, or Ubi-PCNA, were added and temperature was lowered to 25°C. DNA polymerase δ (7 nM) was added after two more min and incubated for 6 min at 25°C. Reactions with PCNA, SUMO-PCNA, and Ubi-PCNA were individually split into Srs2 or buffer only control. (B) Time course of extended D-loop disruption by 7.5 nM Srs2. Samples were taken at 0, 5, 15, and 25 min and stopped by SDS/Proteinase K treatment. (C) Quantitation of total D-loops in (C), unextended D-loops in (D), extended D-loops in (E), and ratio of extended D-loops/unextended D-loops in (F). Plotted are means ± standard deviation from n = 3.
-
Figure 7—source data 1
Source data for Figure 7 and Figure 7—figure supplement 1.
- https://doi.org/10.7554/eLife.22195.025
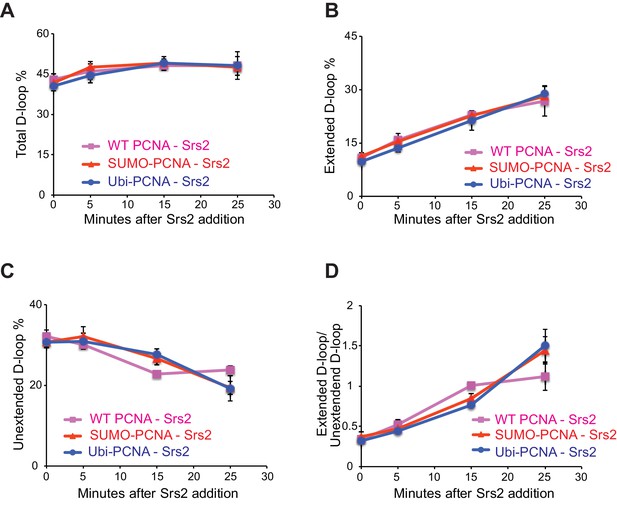
Different species of PCNA, unmodified PCNA, SUMO-PCNA, and Ubi-PCNA, have no impact on the level of unextended D-loops and extended D-loops in the absence of Srs2.
Reactions were described in Figure 6, and quantitation of total D-loops in (A), extended D-loops in (B), unextended D-loops in (C), and ratio of extended D-loops/unextended D-loops in (C). Plotted are means ± standard deviation from n = 3.
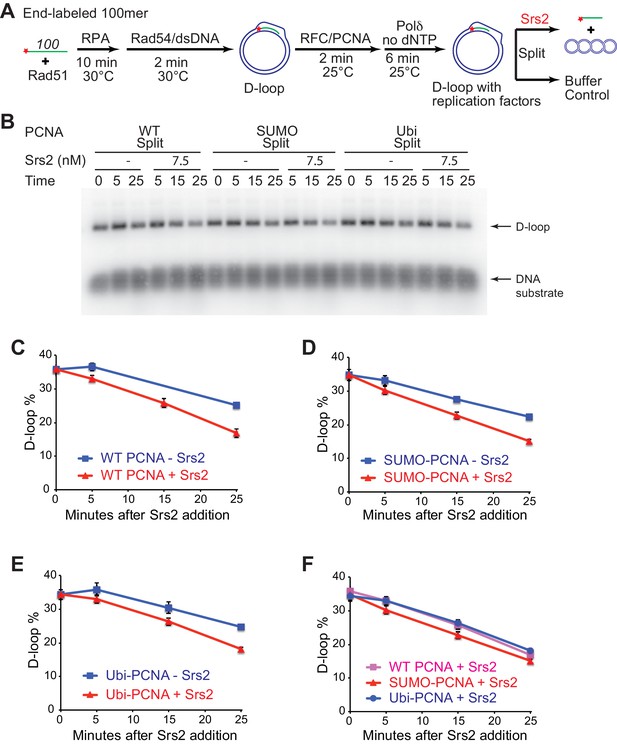
Different species of PCNA, unmodified PCNA, SUMO-PCNA, and Ubi-PCNA, have no impact on the level of D-loops when D-loop extension is blocked.
(A) Extended D-loop disruption assay with RFC/PCNA/Pol δ but without dNTP. Fully homologous 100mer ssDNA was used as substrate. 0.2 µM Rad51 was incubated with 6 nM ssDNA substrates (Rad51/nt = 1:3) for 10 min at 30°C. 165 nM RPA was added for another 10 min incubation before the addition of 84 nM Rad54 (84 nM) and 7 nM supercoiled plasmid dsDNA (21 µM bp). After 2 min, 20 nM RFC and 20 nM PCNA, PCNA-SUMO, or PCNA-Ubi, were added and temperature was switched to 25°C. 7 nM polymerase δ was added after two more min and incubated for six min at 25°C. Reactions with PCNA, PCNA-SUMO, and PCNA-Ubi were individually split into Srs2 or buffer only control. Reaction buffer does not contain dNTP mixture to prevent D-loop extension. (B) Time course of D-loop disruption by Srs2 (7.5 nM), in a reconstituted system with all replication factors but without any dNTP. Samples were taken at 0, 5, 15, and 25 min and stopped by SDS/Proteinase K treatment. (C) Quantitation of D-loops with unmodified PCNA, in the presence or absence of Srs2. (D) Quantitation of D-loops with SUMO-PCNA, in the presence or absence of Srs2. (E) Quantitation of D-loops with Ubi-PCNA, in the presence or absence of Srs2. (F) Comparison of D-loop disruption by Srs2, in the presence of either unmodified PCNA, SUMO-PCNA, or Ubi-PCNA. Plotted are means ± standard deviation from n = 3.
-
Figure 7—figure supplement 2—source data 1
Source data for Figure 7—figure supplement 2.
- https://doi.org/10.7554/eLife.22195.028
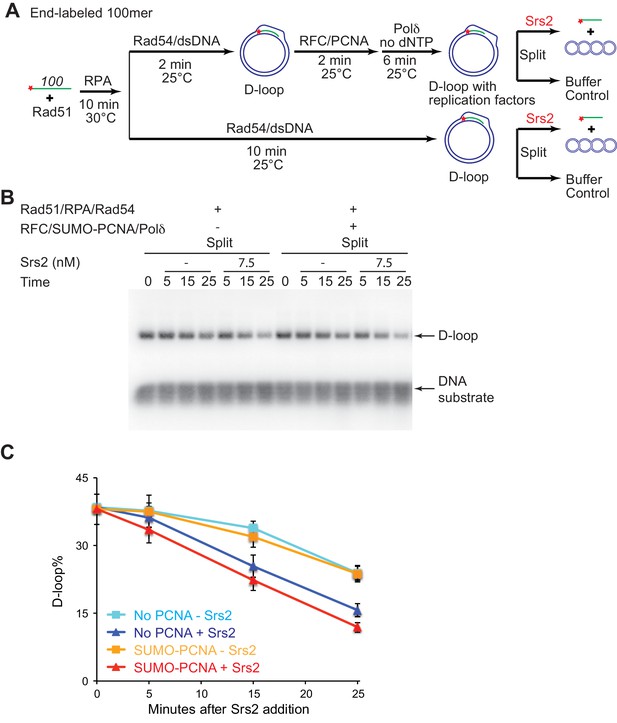
The presence of SUMO-PCNA at the nascent D-loop during Srs2-mediated D-loop disruption.
(A) D-loop disruption assay in the presence or absence of RFC/PCNA/Pol δ and without dNTP. Fully homologous 100mer ssDNA was used as substrates. 0.2 µM Rad51 was incubated with 6 nM ssDNA substrates (Rad51/nt = 1:3) for 10 min at 30°C. 165 nM RPA was added for another 10 min incubation. Then Rad54 (84 nM) and 7 nM supercoiled plasmid dsDNA (21 µM bp) were added and incubated for 2 min at 25°C. The mixture split into buffer containing RFC/PCNA. After two min incubation at 25°C, 7 nM polymerase δ was added into the reaction with RFC/PCNA and incubated for six more min at 25°C. For reactions with or without RFC/PCNA/Pol δ, they were all incubated for 10 min at 25°C in total and then split into Srs2 or buffer only control. The final concentration for RFC is 20 nM; SUMO-PCNA is 20 nM; polymerase δ is 7 nM; Srs2 is 7.5 nM. Reaction buffer does not contain dNTP mixture to prevent D-loop extension. (B) Time course of D-loop disruption by Srs2 in a reconstituted system with all replication factors but without dNTPs. Samples were taken at 0, 5, 15, and 25 min and stopped by SDS/Proteinase K treatment. (C) Quantitation of D-loops in B. Plotted are means ± standard deviation from n = 3.
-
Figure 7—figure supplement 3—source data 1
Source data for Figure 7—figure supplement 3.
- https://doi.org/10.7554/eLife.22195.030
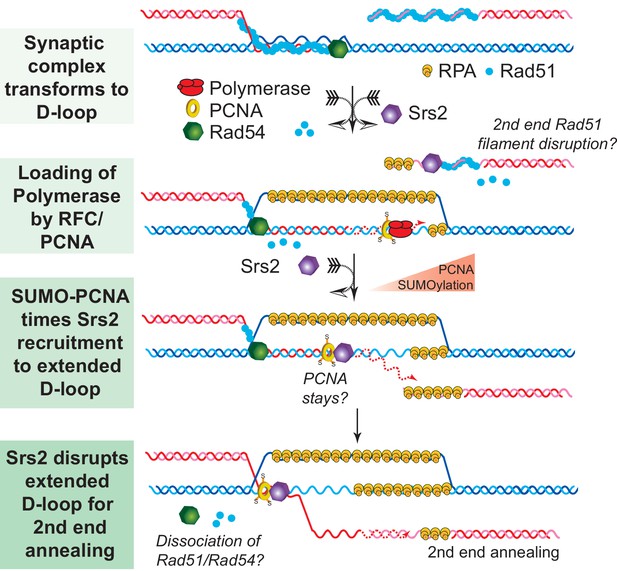
SDSA Model: SUMO-PCNA facilitates Srs2 recruitment towards newly extended D-loops.
After DNA resection and Rad51 filament formation, Rad54 assists Rad51 to search for homology and form D-loops. During heteroduplex formation, Rad54 removes Rad51 at the 3' invading end to prepare the loading of DNA polymerase through RFC and PCNA. DNA synthesis is initiated by DNA polymerase δ. SUMO-PCNA actively recruits Srs2 to the D-loop. Srs2 potentially enforces SDSA through three distinct mechanisms: 1) Srs2 competes with DNA polymerase for PCNA controlling the length of new DNA synthesis (Burkovics et al., 2013). 2) Srs2 translocates in a 3' → 5' direction to dissociate the hDNA incorporated in the D-loop (this work). After the newly extended 3' invading end is dissociated it is available to anneal with the second end to generate non-crossover products. 3) Srs2 may also dissociate Rad51 from the second end to allow single-strand annealing in analogy to RECQ5 (Mitchel et al., 2013; Schwendener et al., 2010).
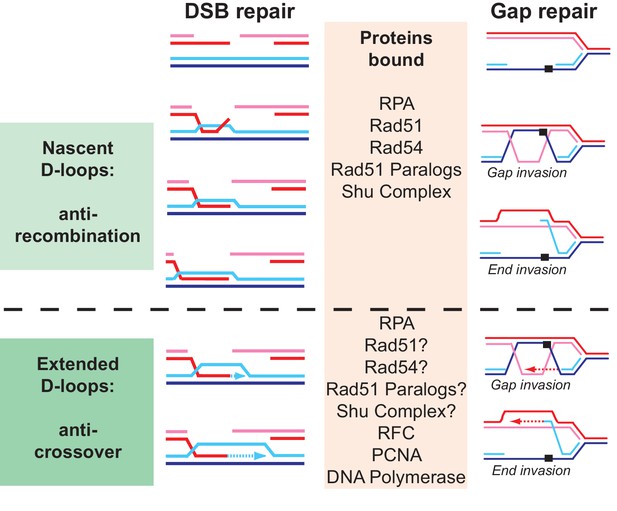
Why are so many proteins involved in D-loop disruption? Different D-loops during homologous recombination initiating at DNA double-stranded breaks and replication-associated gaps.
Numerous proteins have been implicated genetically and biochemically in targeting the D-loop intermediate for disruption as discussed in the introduction. In the budding yeast alone, the DNA helicases Srs2, Sgs1, and Mph1 (Fasching et al., 2015; Marini and Krejci, 2012; Prakash et al., 2009; Sebesta et al., 2011) (this study), the motor protein Rad54 (Bugreev et al., 2007a; Wright and Heyer, 2014), as well as Top3-Rmi1 (Fasching et al., 2015) were shown to disrupt D-loops in vitro. We highlight possible reasons for the involvement of multiple proteins in D-loop disruption. First, before extension by DNA polymerase, disruption of a nascent D-loop is an inherently anti-recombination activity, whereas disruption of a D-loop extended by DNA polymerases limits crossover and favors SDSA. What distinguished these D-loops is their potential length, although little is known about this in vivo, and the proteins bound to the intermediate. While the genetic signatures of anti-recombination and pro-SDSA are different, the biochemical reaction of disrupting nascent or extending D-loops is highly similar and protein interactions involving also posttranslational modifications between the various D-loop disrupting enzymes may only impart preference rather than absolute specificity. Second, D-loops are dynamic entities, likely with varying length and structure, where for example the 3’ end may be embedded in the heteroduplex or transiently extruded. As we show for Srs2 (Figure 4), such structural variations may influence the activity of the enzymes involved. Finally, D-loops occur not only in the context of DSB repair but also in the context of gap repair. The D-loop structure resulting from gap invasion differs, as there are no free ends available to allow true intertwining. We tried to model this reaction using invading DNA molecules with duplex heterology on both ends (Figure 2) and show that Srs2 can disrupt such pairing intermediate, albeit with lower efficiency than regular D-loops. It will be interesting to test the activity of other D-loop disrupting proteins on such substrates. We propose that multiple enzymes with individual preferences for specific D-loop substrates are acting in vivo in a partially overlapping fashion to control anti-recombination, crossover, and SDSA. This view is consistent with genetic data and the observations that mutations in SRS2, SGS1, and MPH1 engage in synthetic lethal/negative interactions in budding yeast (Gangloff et al., 2000; Ira et al., 2003; Mitchel et al., 2013; Prakash et al., 2009).
Tables
Saccharomyces cerevisiae strains used in this study.
| WT | ura3::2x(URA3 Ylplac211 kankanMX4 URA3) | (Ede et al., 2011) |
|---|---|---|
| srs2∆ | ura3::2x(URA3 Ylplac211 kankanMX4 URA3)srs2∆::HisMX6 | This Study |
| srs2-K41A | ura3::2x(URA3 Ylplac211 kankanMX4 URA3)srs2∆::HisMX6 LEU2::srs2-K41A | This Study |
| WDHY 3429 | Wild-type | This Study |
| WDHY 3514 | rad18::LEU2 | Dr. Giordano Liberi (CY8564) |
| WDHY 3858 | srs2∆::KanMX | This Study |
| WDHY 4083 | rad18::LEU2 srs2∆::KanMX | This Study |
| WDHY 3851 | srs2-K41A | Dr. Hanna Klein |
| WDHY 4082 | rad18::LEU2 srs2-K41A | This Study |
| WDHY 4130 | srs2-K41R | Dr. Hanna Klein |
| WDHY 4141 | rad18::LEU2 srs2-K41R | This Study |
| WDHY 5473 | ura3::2x(URA3 Ylplac211 kankanMX4 URA3) | This Study |
| WDHY 5474 | ura3::2x(URA3 Ylplac211 kankanMX4 URA3) rad51::LEU2 | This Study |
| WDHY 5475 | ura3::2x(URA3 Ylplac211 kankanMX4 URA3) srs2::hisMX6 | This Study |
| WDHY 5476 | ura3::2x(URA3 Ylplac211 kankanMX4 URA3) rad51::LEU2 srs2::hisMX6 | This Study |
-
All WDHY strains listed are derivatives of W303 with the common genotype MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 RAD5 (Zhao et al., 1998). All the other strains are derivatives of CEN.PK2-1C (Ede et al., 2011).





