Control of immune ligands by members of a cytomegalovirus gene expansion suppresses natural killer cell activation
Figures
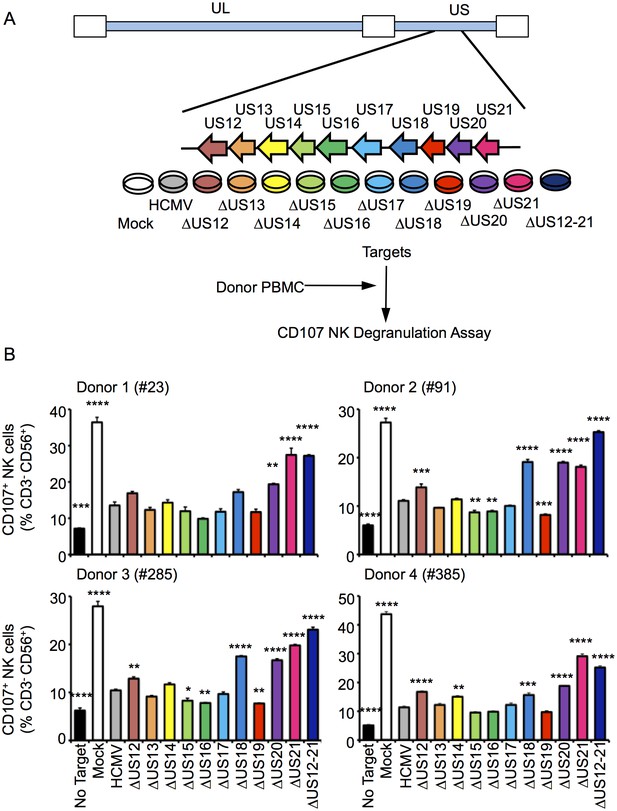
Multiple US12 family proteins regulate NK activation.
(A) Fibroblasts (HF-TERTs) were mock infected or infected with the Merlin strain of HCMV or the series of US12 family deletion mutants for 72 hr. Infected cells were incubated with donor PBMC for 5 hr and NK degranulation assessed by % CD107+ cells within the CD3-, CD56+ population by flow cytometry. (B) CD107 assay results (mean and SD) are shown from four separate donors performed in duplicate or triplicate and analysed by unpaired ordinary one way ANOVA with Dunnett’s test for multiple comparisons against the HCMV control *p<0.05, **p<0.01, ***p<0.005 ***p<0.001). Infected cells were assessed by the % cells with down-regulated MHC I compared to the mock-infected cells (for the experiment using donor #23 and # 385, HCMV 93%, ΔUS12 94%, ΔUS13 98%, ΔUS14 94%, ΔUS15 98%, ΔUS16 99%, ΔUS17 98%, ΔUS18 97%, ΔUS19 99%, ΔUS20 97%, ΔUS21 91%, ΔUS12-21 94%; for the experiment using donor #91 and # 285, HCMV 94%, ΔUS12 94%, ΔUS13 94%, ΔUS14 83%, ΔUS15 95%, ΔUS16 97%, ΔUS17 85%, ΔUS18 94%, ΔUS19 96%, ΔUS20 94%, ΔUS21 94%, ΔUS12-21 96%).
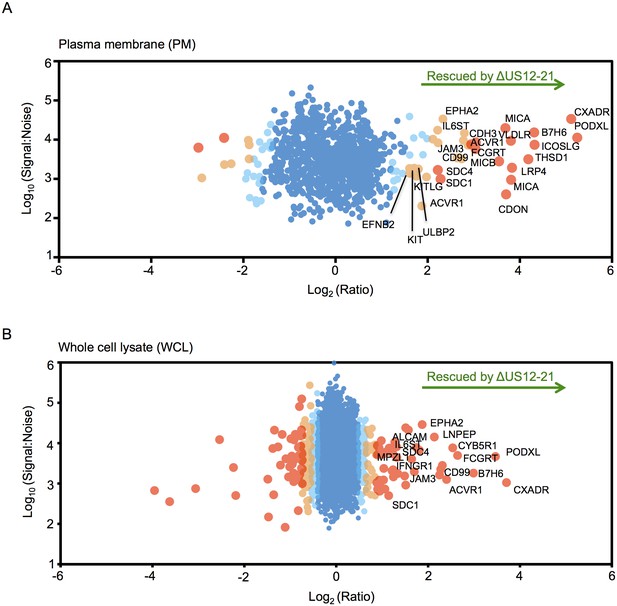
The US12 family targets numerous plasma membrane proteins.
Cells infected with HCMV (Merlin) or HCMV △US12-21 mutant were processed to give PM or WCL fractions and analyzed by TMT mass spectrometry. Scatter plot of proteins identified in the PM (panel A) or WCL fractions (panel B) respectively and quantified by 2 or more unique peptides. Fold change (△US12-21-infected fibroblasts/HCMV-infected cells) is shown as the log2 ratio on the x-axis and the signal:noise on the y-axis as log10. Proteins unaltered by the US21-21 deletion locate at the center of the plots (0 log2/1 fold-change), whereas proteins to the left or right of center represent proteins down- or up-regulated by the US12-21 deletion respectively. Significance B was used to estimate p values (Cox et al., 2009). The 2 different alleles of MICA present in HFs were detected by this analysis.
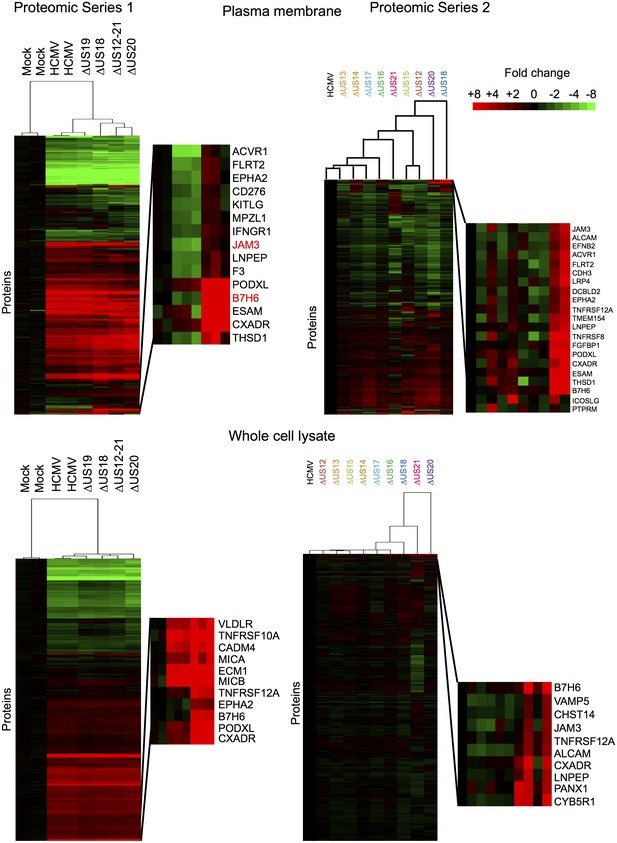
Hierarchical cluster analysis of all proteins quantified in proteomic series 1 and 2.
Zoomed regions are shown for clusters of interest.
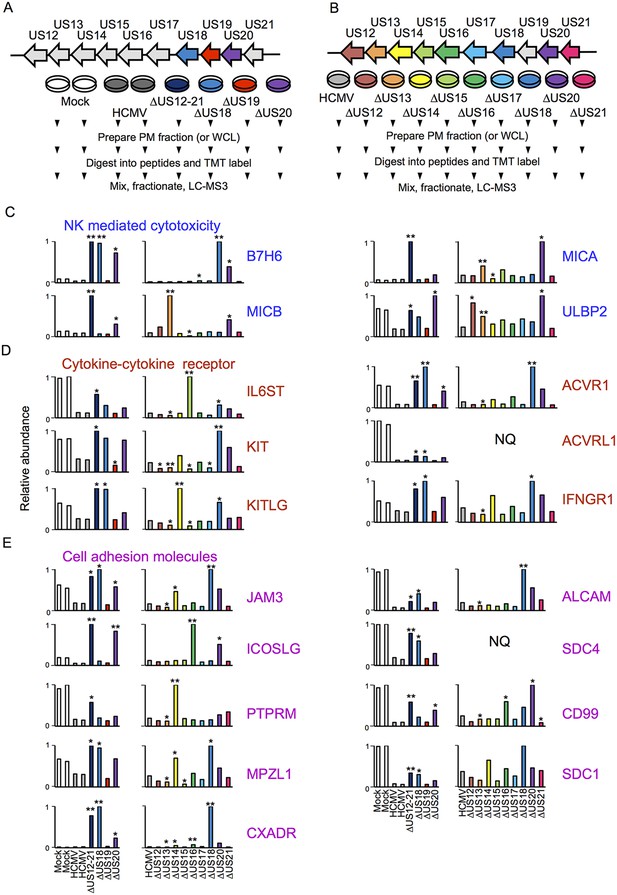
Proteomic analysis of cellular proteins targeted by US12 family members in PM samples.
(A-B) Workflows of proteomic series 1 and 2 respectively. (C-E) Quantitation of PM proteins in enriched KEGG pathways identified using DAVID. Relative abundance of each protein is expressed relative to the sample with the highest abundance (set to 1). NQ – not quantified. A number of the US12-21 mutant targets were also regulated by the US18 and US20 mutants (B7-H6, ULBP2, IL6ST, KIT, KITLG JAM3, ACVR1, ACVRL1, IFNGR1, JAM3, MPZL1, CXADR, ALCAM, SDC4, CD99, SDC1). A subset of these proteins were also regulated by the US14 and/or US16 mutants (KIT, KITLG, ACVR1, IFNGR1, JAM3, MPZL1, CD99, SDC1). We estimated p values for the ratios of each mutant compared to HCMV Merlin using Benjamini-Hochberg corrected Significance B values (Cox and Mann, 2008): *p<0.05, **p<0.0001. For proteomic series 1, ratios were calculated as US12 family deletion mutant / average HCMV and for proteomic series 2, US12 family deletion mutant / HCMV. All proteins quantified by 2 or more peptides were included in this calculation. SDC1 was quantified by a single peptide in proteomic series 2.
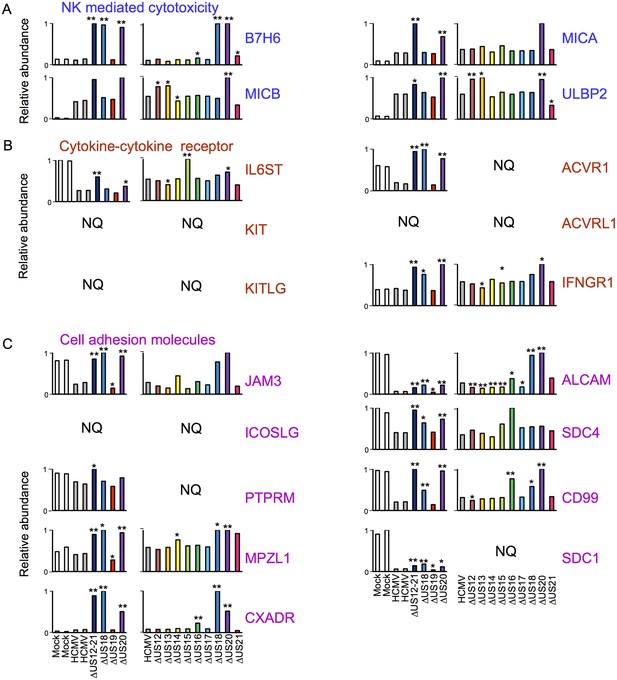
Individual US12-family proteins target key natural killer cell ligands, cell adhesion molecules and cytokines and their receptors in WCL samples.
(A–C) Quantitation of key US12 family targets in WCL samples (proteomic series 1 and 2 - comparative analysis of these proteins from PM samples is shown in Figure 3). We generally observed similar results for proteins quantified in both PM and WCL samples. Relative abundance of each protein is expressed relative to the sample with the highest abundance (set to 1). NQ – not quantified. P values were calculated as described in Figure 4: *p<0.05, **p<0.0001. MICB was only quantified by 1 peptide in proteomic series 1, and MICA, JAM3 and SDC4 by one peptide in proteomic series 2.
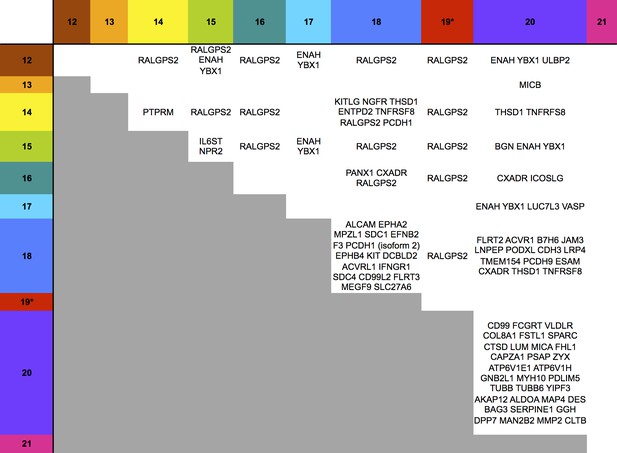
Co-regulation of multiple cell surface proteins by >1 US12 family gene.
Proteins that were rescued by deletion of each US12 family member in the PM fraction of proteomic Series 2 (i.e. not including US19) were analyzed to determine which were additionally targeted by one or more US12 family members. Proteins were plotted in a matrix with unique protein targets on the lower diagonal of the plot (US12 family member compared with itself) and common protein targets in intersections with other US12 members. To identify the highest confidence targets of the US12-21 family for the purposes of this analysis and to generate a shortlist of US12-21 family PM ‘hits’, we employed the following strategy: we included proteins (a) exhibiting at least 3-fold rescue upon deletion of a given US12 family member, quantified by at least two peptides and annotated by GO to indicate a PM location. (b) validated by a corresponding >2 fold change in at least one of (i) deletion of the US12-21 block (proteomic series 1 or 3), (ii) the biological repeat of US18 or US20 in proteomic series 1, (iii) the corresponding gene deletion in WCL proteomic series 2. This strategy identified all proteins shown in Figure 3. *US19 data was used from proteomic series 1.
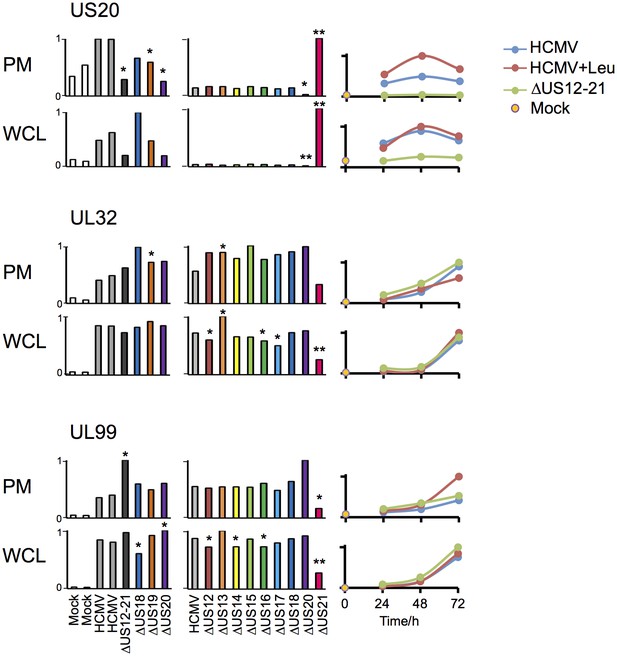
Altered expression of US20, UL32 and UL99 levels in the US21 deletion mutant-infected fibroblasts.
Quantitation of US20, UL32 and UL99 in proteomic series 1–3 in both PM and WCL. Abundance of each protein is expressed relative to the sample with the highest abundance (set to 1). For proteomic series 1 and 2, p values were calculated as described in Figure 4: *p<0.05, **p<0.0001. US20 was only quantified by one peptide in proteomic series 1, WCL experiment.
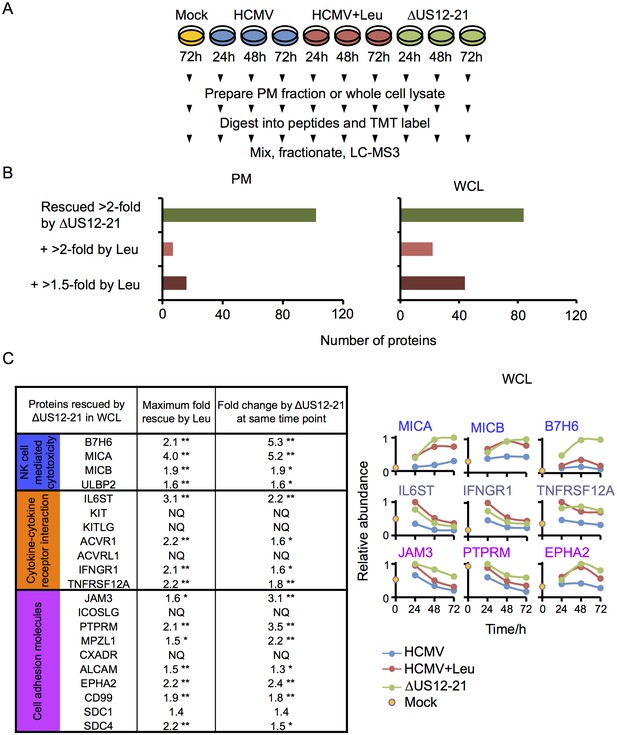
Multiple US12 gene family targets are degraded via the lysosomal pathway.
(A) Workflow of Proteomic Series 3. (B) Number of proteins targeted by the US12-21 block and additionally rescued >2 fold in the dataset by the △US12-21 deletion mutant or rescued >2 fold or >1.5 fold by leupeptin treatment for both WCL and PM. C. Comparable degree of rescue of US12-21 target proteins by US12-21 deletion and leupeptin treatment. Quantitation of a subset of these proteins is shown relative to the maximum abundance (set to 1). p-values were calculated as described in Figure 4, for the ratios of leupeptin treatment or US12-21 deletion virus infection at each time point compared to the matched wild-type Merlin-infected control. *p<0.05, **p<0.0001. SDC1 was only quantified by one peptide.
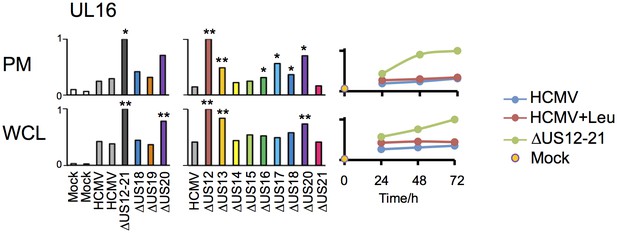
Regulation of UL16 levels by the US12 family.
Quantitation of UL16 in proteomic series 1–3 in both PM and WCL. Relative abundance of each protein is expressed relative to the sample with the highest abundance (set to 1). For proteomic series 1 and 2, p values were calculated as described in Figure 4: *p<0.05, **p<0.0001.
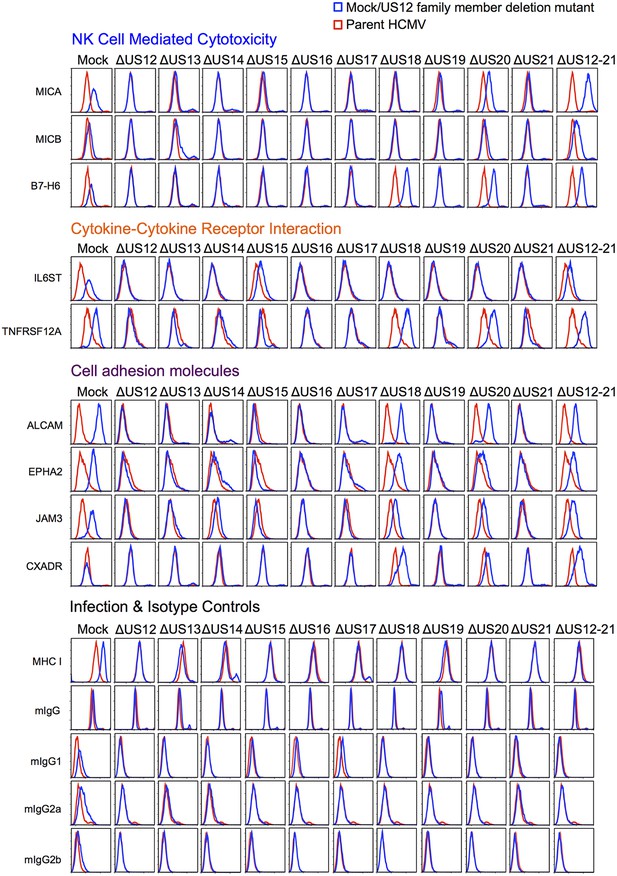
Validation of cell surface proteins regulated by the US12 family.
Flow cytometry confirmed proteomic data for proteins representative of each category enriched in the DAVID analysis. Staining in mock/US12 family member deletion mutant infections (blue line) is shown relative to the parental HCMV infection (red line). Flow cytometry was carried out for cell surface expression of MHC I (W6/32) as a control for HCMC infection and isotype antibody staining controls (with directly PE-conjugated IgG1, IgG2a or IgG2b antibodies or for unconjugated antibodies mIgG and an anti-mouse-AF647 conjugated secondary antibody). Infected cells were assessed by the % cells with down-regulated MHC I compared to the mock-infected cells (HCMV 94%, ΔUS12 94%, ΔUS13 94%, ΔUS14 83%, ΔUS15 95%, ΔUS16 97%, ΔUS17 85%, ΔUS18 94%, ΔUS19 96%, ΔUS20 94%, ΔUS21 94%, ΔUS12-21 96%). Results are representative of at least two independent experiments.
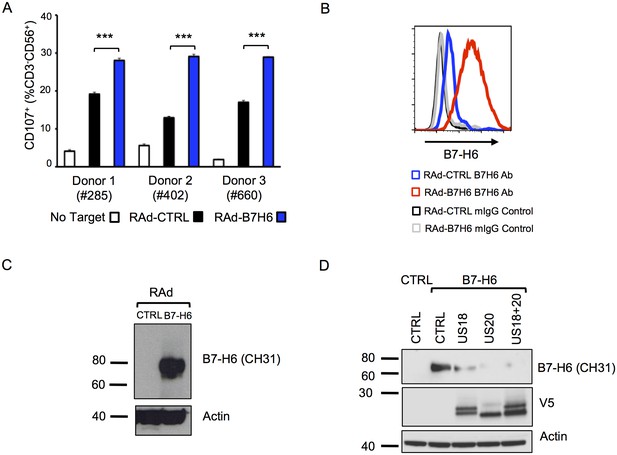
Adenovirus expressed B7-H6 regulates NK cell activation and B7-H6 levels are regulated by ectopically-expressed US18 and US20.
(A) HF-CARs were infected with control (RAd-CTRL) or B7-H6-expressing (RAd-B7-H6) adenovirus vectors (MOI 5). Cells were harvested 48 h p.i. and used as targets in a CD107 degranulation assay with buffy-coat derived PBMC from 3 separate donors in duplicate or triplicate. Results (shown as mean and SD) were analyzed by unpaired two-tailed Student’s T-test. ***p<0.001, ****p<0.0001. (B and C) HF-CARs were infected with control (RAd-CTRL) or B7-H6-expressing (RAd-B7-H6) adenovirus vectors (MOI 5). Cells were harvested 48 h p.i. and B7-H6 cell surface expression analyzed by flow cytometry (B) or western blot (C). Results are representative of at least 2 independent experiments. (D) HF-CARs were infected with adenovirus control (CTRL), US18-expressing adenovirus, US20-expressing adenovirus or a combination of both US18 and US20 (MOI 5 each, made up to a total MOI of 10 with RAd-CTRL). After 24 hr incubation, HF-CARs were infected with adenovirus control (CTRL) or B7-H6-expressing adenovirus (B7–H6) at MOI 5 as indicated before incubation for a further 48 hr. Whole cell lysates were prepared and analysed by immunoblotting with the antibodies indicated. Results are representative of two independent experiments.
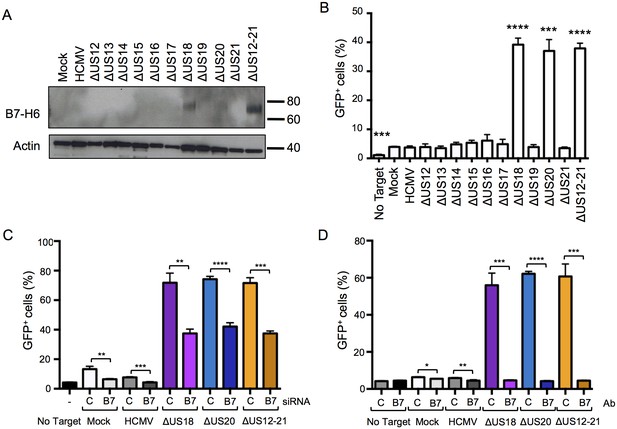
Differences in B7-H6 levels on HCMV US12 family deletion mutant-infected cells regulate NKp30-mediated responses.
(A) Whole cell lysates were prepared from mock, HCMV or the series of US12 family deletion mutant infected fibroblasts and analyzed by immunoblotting with antibodies specific for B7-H6 (CH31) or actin. Results are representative of two independent experiments. (B) An NKp30-responsive 2B4 reporter cell line containing a NFAT-GFP reporter construct (CT299) was incubated for 24 hr with mock, HCMV or the series of US12 family deletion mutant infected fibroblasts in triplicate. Fixed cells were then analyzed for GFP fluorescence by flow cytometry compared to cells incubated with no target fibroblasts. Results are expressed as the % GFP positive reporter cells (mean and SD) and were analyzed by unpaired two-tailed Student’s T-test. Results are representative of two independent experiments. (C) HF-TERTs were transfected with control (C) or B7-H6 (B7) siRNAs for 24 hr prior to infection with the parent HCMV, △US18, △US20 or △US12-21 mutants for 72 hr. Cells were then incubated for 24 hr with CT299 NKp30 reporter cells in triplicate and GFP+ cells determined by flow cytometry, and analysed by unpaired two-tailed Student’s t-test. Results (mean and SD) are representative of two independent experiments. (D) HF-TERTs infected with HCMV, △US18, △US20 or △US12-21 mutants were incubated for 24 hr with CT299 NKp30 reporter cells, in the presence isotype control (C) or B7-H6 blocking antibodies (B7) in triplicate, GFP+ cells determined by flow cytometry and analyzed by unpaired two-tailed Student’s t-test. Results (mean and SD) are representative of two independent experiments.
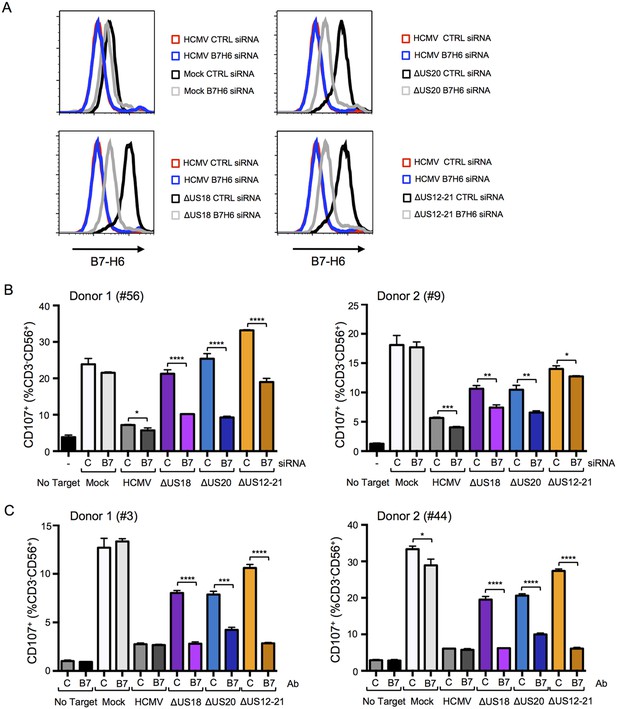
B7-H6 regulation has a major effect on NK activation in response to HCMV infection.
(A and B) HF-TERTs were transfected with control (C) or B7-H6 (B7) siRNAs for 24 hr prior to infection with the parent HCMV, △US18, △US20 or △US12-21 mutants. Cells were harvested 72 h p.i. and B7-H6 cell surface expression analyzed by flow cytometry (E) or used as targets in a CD107 degranulation assay with donor-derived PBMC in triplicate (F). Results (mean and SD) were analyzed by unpaired two-tailed Student’s t-test and are representative of two independent experiments using 2 separate donors in each. Infected cells were assessed by the % cells with down-regulated MHC I compared to the mock-infected cells (HCMV CTRL siRNA 95%, ΔUS18 CTRL siRNA 91%, ΔUS20 CTRL siRNA 93%, ΔUS12-21 CTRL siRNA 86%, HCMV B7-H6 siRNA 96%, ΔUS18 B7-H6 siRNA 84%, ΔUS20 B7-H6 siRNA 96%, ΔUS12-21 B7-H6 siRNA 79%). (C) HF-TERTs infected with HCMV, △US18, △US20 or △US12-21 mutants were used as targets in a CD107 degranulation assay with donor-derived PBMC, in the presence isotype control (C) or B7-H6 (B7) blocking antibodies in triplicate. Results (mean and SD) were analyzed by unpaired two-tailed Student’s t-test and are shown for two separate donors. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Tables
HCMV constructs used in the study.
| Virus | BAC # | Accession no. | Cassette used | Modification | Previous reference |
|---|---|---|---|---|---|
| HCMV | 1111 | GU179001.1 | None | RL13-, UL128- | Stanton et al. (2010) |
| △US12 | 1810 | KU221097 | GalK | US12 CDS deleted | None |
| △US13 | 1831 | KU221099 | GalK | US13 CDS deleted | None |
| △US14 | 1798 | KU221093 | GalK | US14 CDS deleted | None |
| △US15 | 1800 | KU221094 | GalK | US15 CDS deleted | None |
| △US16 | 1802 | KU221095 | GalK | US16 CDS deleted | None |
| △US17 | 1804 | KU221096 | GalK | US17 CDS deleted | None |
| △US18 | 1654 | KU221091 | RpsL-Neo-LacZ | US18 CDS deleted | Fielding et al. (2014) |
| △US19 | 1796 | KU221092 | RpsL-Neo-LacZ | US19 CDS deleted | None |
| △US20 | 1595 | KU221090 | SacB-AmpR-LacZ | US20 CDS deleted | Fielding et al. (2014) |
| △US21 | 1871 | KU221100 | RpsL-Neo-LacZ | US21 CDS deleted | None |
| △US12-21 | 1815 | KU221098 | RpsL-Neo-LacZ | US12-21 deleted | None |
Primers used in the construction of the HCMV US12 deletion mutants library.
| Primer | Sequence (5’ > 3’) |
|---|---|
| US12 GalK For | GCGGGGGACAAAGGACAGTACGACAGATTAGGTGATAGAAACGTTTTTTTCCTGTTGACAATTAATCATCGGCA |
| US12 GalK Rev | AAACTTGCCGGGTACCTGAAGCCCCGACGACTGTTCGTCGAGCACCCG TCTCAGCACTGTCCTGCTCCTT |
| US12 Delete | GCGGGGGACAAAGGACAGTACGACAGATTAGGTGATAGAAACGTTTTTTTGACGGGTGCTCGACGAACAGTCGTCGGGGCTTCAGGTACCCGGCAAGTTT |
| US13 GalK For | CTTCAGGTACCCGGCAAGTTTTATAGAGAAAGGGGGACGATGGGTGGTGGCCTGTTGACAATTAATCATCGGCA |
| US13 GalK Rev | GAAGACTCCACCGAGACGCTCACCCGTTCACTCGGGCGCATCACCCGC CTTCAGCACTGTCCTGCTCCTT |
| US13 Delete | CTTCAGGTACCCGGCAAGTTTTATAGAGAAAGGGGGACGATGGGTGGTGGAGGCGGGTGATGCGCCCGAGTGAACGGGTGAGCGTCTCGGTGGAGTCTTC |
| US14 GalK For | GAGTGAACGGGTGAGCGTCTCGGTGGAGTCTTCTTATAAACCAGCGGG TCCCTGTTGACAATTAATCATCGGCA |
| US14 GalK Rev | CTGTAGCTTCGAGACCTTGCGGATACGCCGCCGGGCGCTGCGGTCCCG ACTCAGCACTGTCCTGCTCCTT |
| US14 Delete | GAGTGAACGGGTGAGCGTCTCGGTGGAGTCTTCTTATAAACCAGCGGGTCGTCGGGACCGCAGCGCCCGGCGGCGTATCCGCAAGGTCTCGAAGCTACAG |
| US15 GalK For | CTCCATGTCGGGACCGCAGCGCCCGGCGGCGTATCCGCAAGGTCTCGAAGCCTGTTGACAATTAATCATCGGCA |
| US15 GalK Rev | CGGAACTGGTTTTCGGACAGAGCAGCCGTTTCCAGAGAACGCAGCGCA CCTCAGCACTGTCCTGCTCCTT |
| US15 Delete | CTCCATGTCGGGACCGCAGCGCCCGGCGGCGTATCCGCAAGGTCTCGAAGGGTGCGCTGCGTTCTCTGGAAACGGCTGCTCTGTCCGAAAACCAGTTCCG |
| US16 GalK For | CGTTCTCTGGAAACGGCTGCTCTGTCCGAAAACCAGTTCCGAACGAAAATCCTGTTGACAATTAATCATCGGCA |
| US16 GalK Rev | CCCCACGGATCTCGCGTCTTAGACGCGCGGTCATATAGCCTCCGGCTG TCTCAGCACTGTCCTGCTCCTT |
| US16 Delete | CGTTCTCTGGAAACGGCTGCTCTGTCCGAAAACCAGTTCCGAACGAAAATGACAGCCGGAGGCTATATGACCGCGCGTCTAAGACGCGAGATCCGTGGGG |
| US17 GalK For | TTGGTGGAGACGGCCGGCGCGGCGGGTGGGGGAAACGACGAGTTTTTCCGCCTGTTGACAATTAATCATCGGCA |
| US17 GalK Rev | ACACTCTATAAACGGTTTCTCATACGCGCCTTTTGATAGCCACCGCCG TCTCAGCACTGTCCTGCTCCTT |
| US17 Delete | TTGGTGGAGACGGCCGGCGCGGCGGGTGGGGGAAACGACGAGTTTTTCCGGACGGCGGTGGCTATCAAAAGGCGCGTATGAGAAACCGTTTATAGAGTGT |
| US19 SacB For | CAGCACCCGGTTACCGCGGATTTGATTGACGTCACGAGTGTGGTCAAACCGTGGCGGCACCCTGTATCCGACCCGTCGCCTGTGACGGAAGATCACTTCG |
| US19 SacB Rev | GCTACGCCTCTATGTCGAAAATGTGGCTTTATTCATCGGCATGTACCATCTTCTGAGGCTCTGGTTGTGGAGCCCATGACTGAGGTTCTTATGGCTCTTG |
| US19 Delete | ACGTCACGAGTGTGGTCAAACCGTGGCGGCACCCTGTATCCGACCCGTCGGGCGACAAGCGCGGCTGCTGTGAAAACGGGCGCGGTTTTATAGGCATTAG |
| US21 SacB For | TGCGGCGCACCTACCCTTCTCTTATACACAAGCGAGCGAGTGGGGCACGGTGACGTGGTCACGCCGCGGACACGTCGACCTGTGACGGAAGATCACTTCG |
| US21 SacB Rev | CAGCGCCCACACTGCTCAGACGACGGTCGCTGCGACGGTCGCTGCCACAGCAGCGGCGTCGCCCCAGTTCGTCTCCTAACTGAGGTTCTTATGGCTCTTG |
| US21 Delete | CAAGCGAGCGAGTGGGGCACGGTGACGTGGTCACGCCGCGGACACGTCGAGGCGGCAACGCCGGCGGTTATCGCCGAGATTCGTCTAAATACACGAAGCG |
| US12-21 Delete | GCGGGGGACAAAGGACAGTACGACAGATTAGGTGATAGAAACGTTTTTTTGGCGGCAACGCCGGCGGTTATCGCCGAGATTCGTCTAAATACACGAAGCG |
Primers used in the local sequencing of the HCMV US12 deletion mutant library
| Primer | Sequence (5’ > 3’) |
|---|---|
| US12 Seq For | CCCTGTCTAGACTCAAAAGCTG |
| US12 Seq Rev | ATCGTCCCCCTTTCTCTATA |
| US13 Seq For | GCCGAGTGGCTCGCC |
| US13 Seq Rev | CTGGGCACCTATCATCATTA |
| US14 Seq For | GGAGGGAAGCCCATTGC |
| US14 Seq Rev | TCATTACCTGTCTAGCCG |
| US15 Seq For | CGGACGCGGCTTCC |
| US15 Seq Rev | GTCGCTACAGCTCTTTATTA |
| US16 Seq For | GGGGCACGTAGATGACCG |
| US16 Seq Rev | CTCATTAGACAAACTCATCG |
| US17 Seq For | GTCTAAGACGCGAGATCCG |
| US17 Seq Rev | CCCAGTAGACAGACAGAACA |
| US19 Seq For | GGAGCGGCACGATGGTGACC |
| US19 Seq Rev | TCTGCCCACCTAACCAATGC |
| US21 Seq For | GCTGAAAGATGAAGATGGCG |
| US21 Seq Rev | ACCCGACCAGATGGGAGACG |
Additional files
-
Supplementary file 1
Interactive spreadsheet of all US12 family proteomic data (separate Excel Spreadsheet).
This spreadsheet enables the generation of graphs showing the relative expression of any of the human and HCMV proteins quantified across Proteomic series 1–3 in PM and WCL fractions by typing the gene name into the indicated box. The datasets used to generate the graph and protein aliases are present in the other tabs.
- https://doi.org/10.7554/eLife.22206.019





