Nucleosomes influence multiple steps during replication initiation
Figures
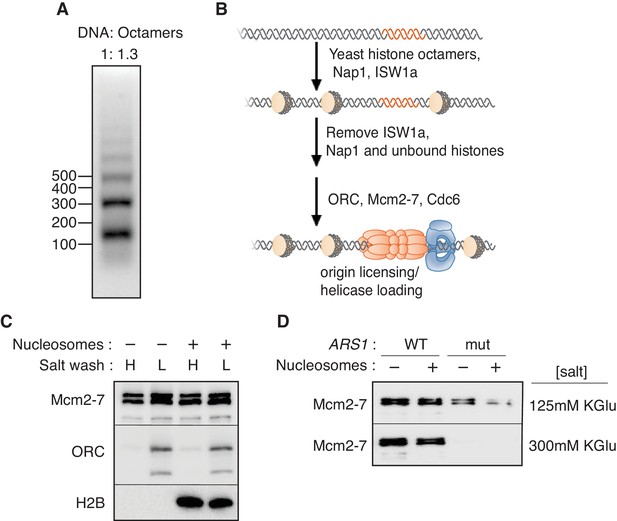
Mcm2-7 helicase loading onto nucleosomal DNA templates.
(A) Nucleosomes were remodeled with bead-coupled ARS1-containing linear DNA, ISW1a, yeast histone octamers and Nap1. Nucleosome assembly was assessed after partial MNase digestion. (B) Outline of the helicase-loading assay using nucleosomal DNA. (C). Comparison of helicase loading on naked DNA and on ISW1a-remodeled nucleosomal DNA. DNA templates were washed with high-salt (H) or low-salt (L) buffer after loading. Template-associated Mcm2-7, ORC and H2B was detected by immunoblot. (D) Helicase loading onto either wild-type (WT) or A-B2- (mut) (Heller et al., 2011) ARS1-containing DNA. As indicated, nucleosomal DNA was remodeled with ISW1a. Assays were performed in either 125 mM (to allow increased origin non-specific helicase loading) or 300 mM (origin specific helicase loading) potassium glutamate. After a high salt wash, DNA-associated Mcm2-7 was detected by immunoblot.
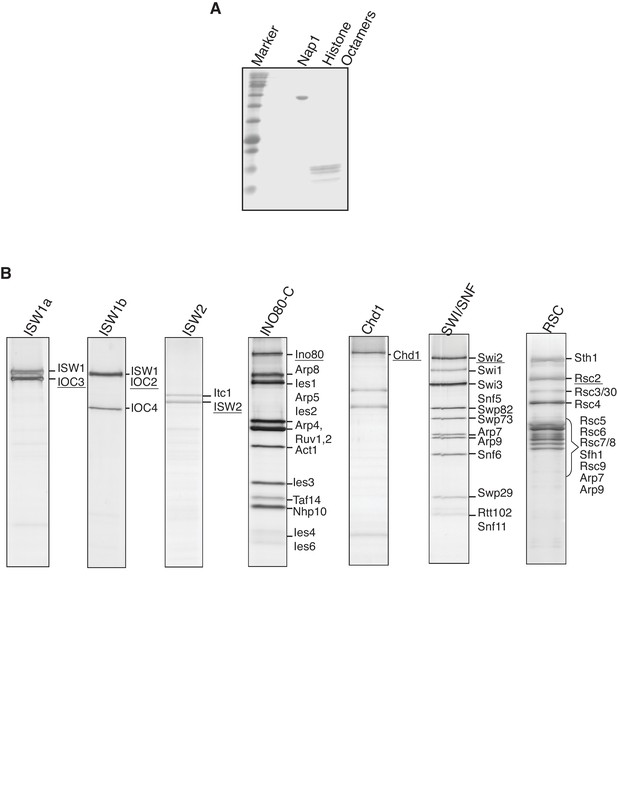
Purified proteins used in the in vitro nucleosome.
assembly reactions. (A) Purified yeast histone octamers and Nap1 were separated by. SDS-AGE and visualized by Coomassie staining. (B) Purified CREs were separated. by SDS-PAGE and visualized by Coomassie staining.
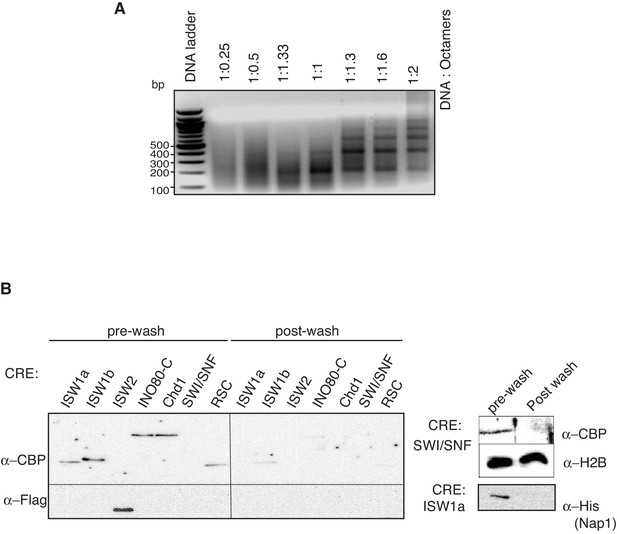
Preparation of in vitro nucleosome templates.
(A) Nucleosomes were assembled with increasing amounts of histone octamers and fixed amount of DNA with ISW1a. Nucleosome assembly was as in Figure 1A. (B) Removal of CRE and unassociated proteins from in- vitro assembled nucleosomes. Amount of CRE and Nap1 associated with nucleosomal DNA before and after washing was detected by anti-CBP (Ioc3-TAP, Ioc2-TAP, Ino80-TAP, Chd1-TAP, Swi2-TAP and Rsc2-TAP), anti-FLAG (Isw2-FLAG), anti-H2B and anti-6xHis (Nap1) immunoblots.
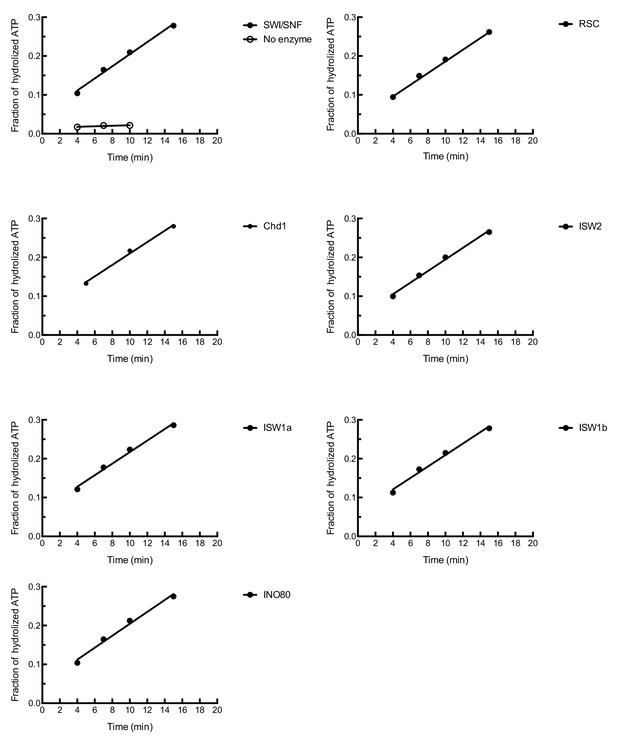
The ATPase activities of in vitro purified chromatin remodeling enzymes.
The ATPase activities of chromatin remodeling enzymes were measured in the presence of 0.1 mg/ml plasmid DNA. The fractions of hydrolyzed ATP were normalized with 1 nM remodeling enzymes.
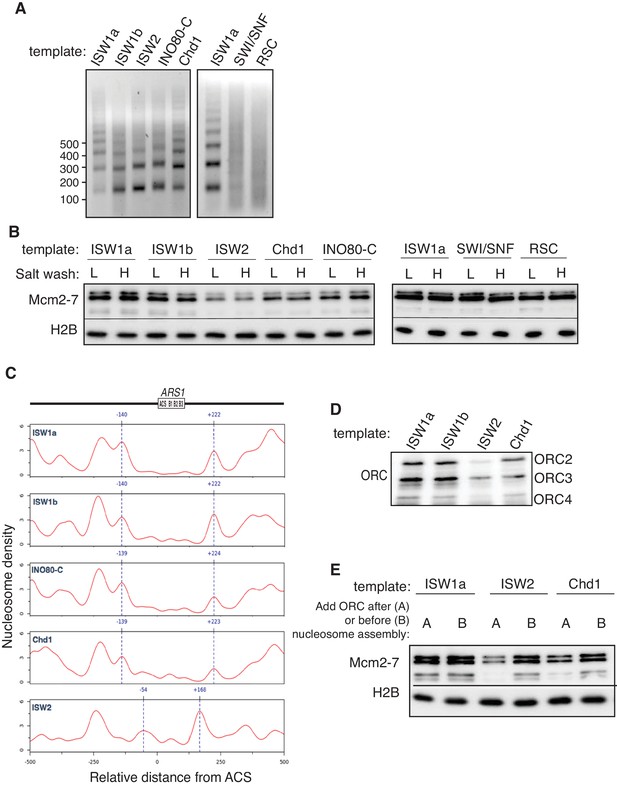
Comparison of helicase loading onto nucleosomal DNA templates remodeled with different CREs.
(A) Comparison of nucleosome assembly with different CREs. Nucleosomes were remodeled with the indicated CRE and assayed by partial MNase digestion. (B) Helicase loading onto nucleosomes remodeled with different CREs. After helicase loading, DNA was washed either with high-salt (H) or low-salt (L) buffer. Mcm2-7 and H2B DNA association was detected by immunoblot. (C) Comparison of origin-proximal nucleosome positioning established by different CREs. The positions of nucleosome dyads remodeled with the indicated CRE were analyzed by high-throughput MNase-Seq. Nucleosome dyad density (Y-axis) and the corresponding position of the dyad (X-axis) are plotted. Zero on the X-axis indicates the first nucleotide of the ARS1 consensus sequence (ACS). The elements of ARS1 (Marahrens and Stillman, 1992) are indicated above. (D) ORC association with nucleosomal DNA remodeled with different CREs. Template association of ORC was detected by immunoblot. (E) Addition of ORC during nucleosome assembly restores helicase loading on ISW2 and Chd1 templates. Nucleosomes were assembled onto ARS1 DNA with the indicated CRE in the presence or absence of ORC. Helicase loading was performed and analyzed as described in (B).
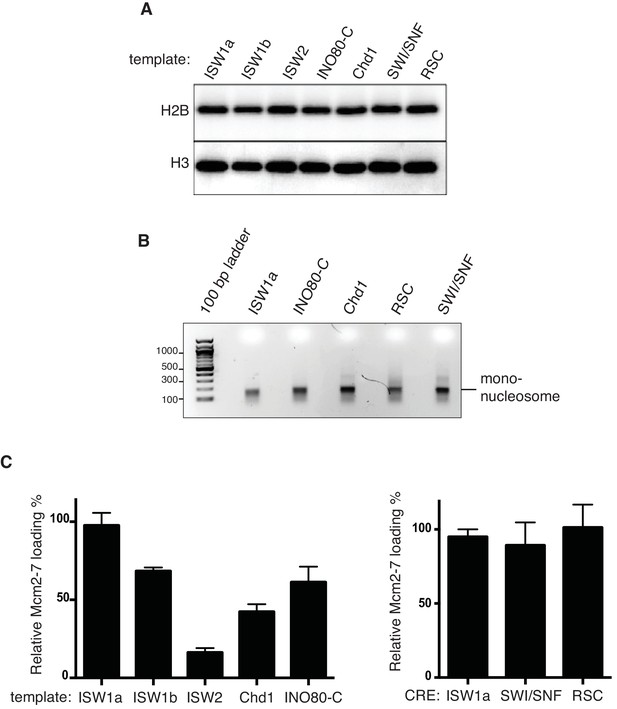
Nucleosome assembly with different CREs and their ability to load Mcm2-7 helicase.
(A) Histone H2B and H3 associated with nucleosomes assembled with the indicated CREs was detected by anti-H2B and H3 immunoblot. (B) Mono-nucleosomes produced from nucleosomal templates assembled with different CREs Similar amounts of ISW1a-, INO80-C, Chd1-, RSC- and SWI/SNF assembled templates were digested extensively with MNase and purified mononucleosomal DNA was analyzed by agarose gel electrophoresis. (C) Quantification of relative Mcm2-7 loading for nucleosomal templates assembled with the indicated CRE. The amount of Mcm2-7 quantified using online software ImajeJ and statistical analysis was performed using Prism software. For each assay three (n = 3) biological replicates were quantified. Mean value for ISW1a (High salt wash) reactions was calculated and set as the 100%. All the other values were calculated as a percentage of that mean value. Error bars indicate standard deviation (SD).
-
Figure 2—figure supplement 1—source data 1
Raw values used in the quantification of Figure 2B, left panel (n = 3).
- https://doi.org/10.7554/eLife.22512.009
-
Figure 2—figure supplement 1—source data 2
Raw values used in the quantification of Figure 2B, right panel (n = 3).
- https://doi.org/10.7554/eLife.22512.010
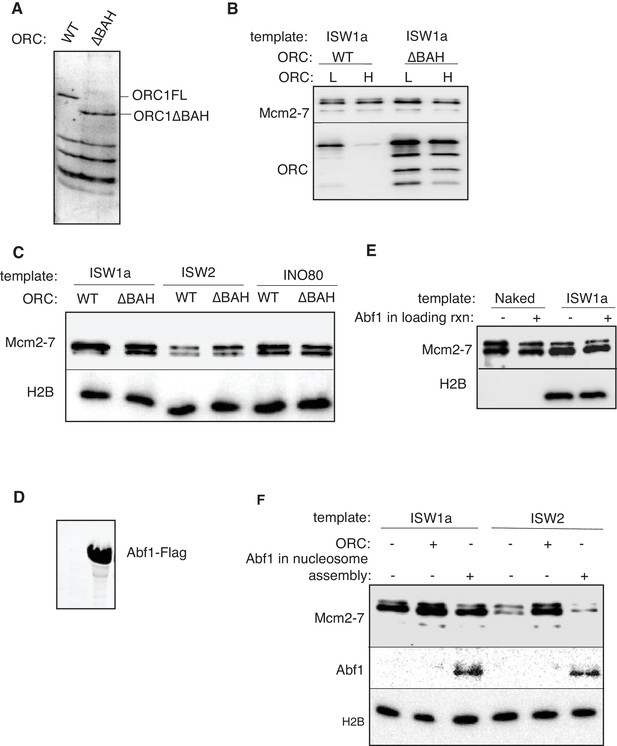
ORC1 BAH domain and Abf1 is dispensable for helicase loading of nucleosomal templates.
(A) Purified ORC and ORCΔBAH were separated by SDS-PAGE and visualized by Coomassie staining. (B) Comparison of WT. and ΔBAH ORC mediated helicase loading on ISW1a-remodeled DNA templates. DNA templates were washed with high-salt (H) or low-salt (L) buffer after loading. Template associated Mcm2-7 and ORC was detected by immunoblot. (C) Helicase loading onto ISW1a, ISW2 and INO80-C templates were carried out with WT or ΔBAHORC. DNA templates were washed with high-salt (H) after loading. Template-associated Mcm2-7 and H2B was detected by immunoblot. (D) Purified Abf1 was separated by SDS-PAGE and visualized by Coomassie staining. (E) Comparison of Mcm2-7 loading for naked DNA and ISW1aremodeled nucleosomal templates. After helicase loading, DNA was washed with high salt. Mcm2-7 and H2B DNA association was detected by immunoblot. (F) Addition of Abf1 during nucleosome assembly do not restore helicase loading defects of ISW2 templates. Nucleosomes were remodeled onto ARS1 DNA with the indicated CRE in the presence/absence of ORC or Abf1 and ability load helicase was determined. Template associated Mcm2-7, Abf1(anti-Flag) and H2B was detected by immunoblot after high salt wash.
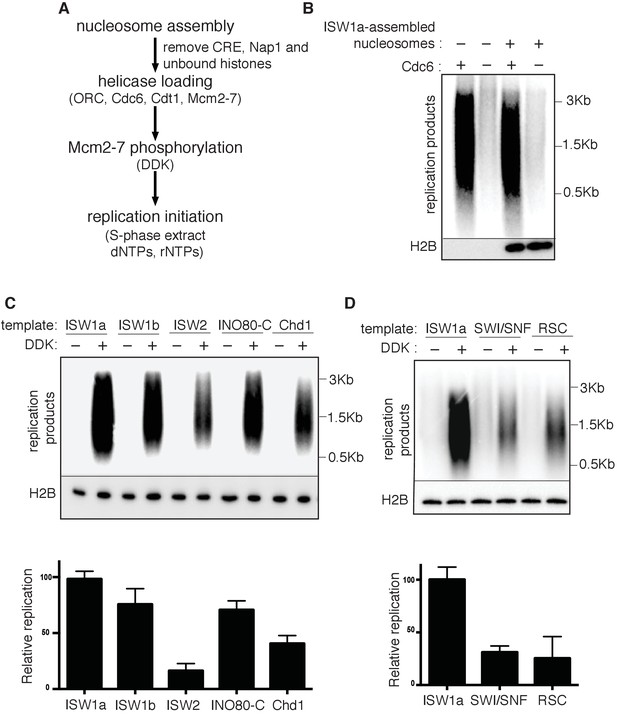
Replication initiation on nucleosome templates.
(A) Outline of nucleosomal DNA replication initiation assay using purified proteins and yeast S-phase cell extract. (B) ISW1a templates do not interfere with replication. Naked DNA or ISW1a templates were assayed in the presence or absence of Cdc6. Radiolabeled replication products were analyzed by alkaline agarose electrophoresis and autoradiography (top). Template-associated H2B was detected by immunoblot (lower). (C) Comparison of replication using ISW1a, ISW1b, ISW2, INO80-C and Chd1 templates in the presence and absence of DDK. Products of the extract-based replication assays were analyzed as in (B, top). H2B levels for each template are shown (middle). Quantification of replication products was performed as in Figure 2B. Error bars show the SD (n = 3, lower). (D) Comparison of replication of ISW1a, SWI/SNF and RSC templates in the presence or absence of DDK. Analysis of replication products, template-associated H2B and quantification (n = 3) as in (C).
-
Figure 3—source data 1
Raw values used in the quantification of Figure 3C (n = 3).
- https://doi.org/10.7554/eLife.22512.013
-
Figure 3—source data 2
Raw values used in the quantification of Figure 3D (n = 3).
- https://doi.org/10.7554/eLife.22512.014
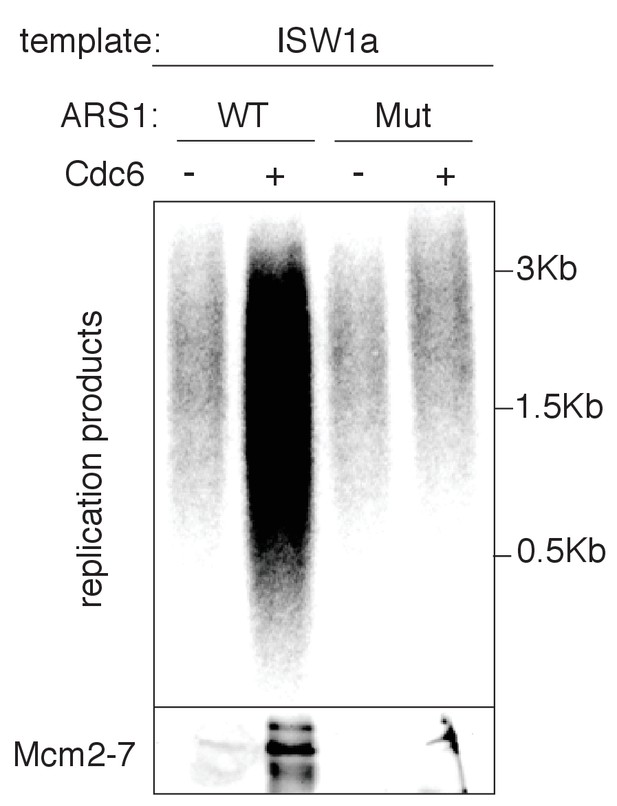
In vitro nucleosomal DNA template replication initiation is origin specific.
WT and mutant ARS (A-B2-) containing ISW1a templates were assayed in the presence or absence of Cdc6. Radiolabeled replication products were analyzed by alkaline agarose electrophoresis and autoradiography (top). Template-associated Mcm2-7 was detected by immunoblot (lower). Products of the extract-based replication assay were analyzed as in Figure 3B.
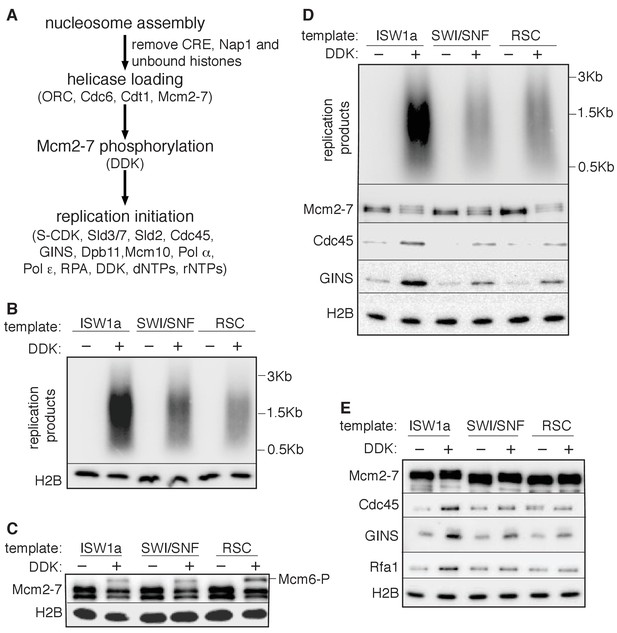
SWI/SNF and RSC templates show reduced CMG formation.
(A) Outline of fully-reconstituted nucleosomal DNA replication initiation assay. The proteins added at each step are indicated. (B) Comparison of reconstituted nucleosomal DNA replication using ISW1a, SWI/SNF and RSC templates in the presence or absence of DDK. Analysis of replication products and H2B as in Figure 3B. (C) Comparison of Mcm2-7 phosphorylation by DDK on ISW1a, SWI/SNF and RSC templates. Phosphorylation of Mcm6 is indicated by reduced electrophoretic mobility and was analyzed by immunoblot (top). Template associated H2B is shown (lower). (D) Comparison of replication of ISW1a, RSC and SWI/SNF templates. Reactions were performed with or without DDK and replication products of the reconstituted replication reactions were analyzed as in Figure 3B (top). Template association of Mcm2-7, Cdc45, GINS and H2B was measured after a high-salt wash at the end of reconstituted replication assay by immunoblot (lower panels). (E) Comparison of CMG formation and activation using ISW1a, SWI/SNF and RSC templates. To prevent replication initiation, the only nucleotide present was ATP and Pol α was left out of the assay. Template association of Mcm2-7, Cdc45, GINS, Rfa1 and H2B were measured by immunoblot.
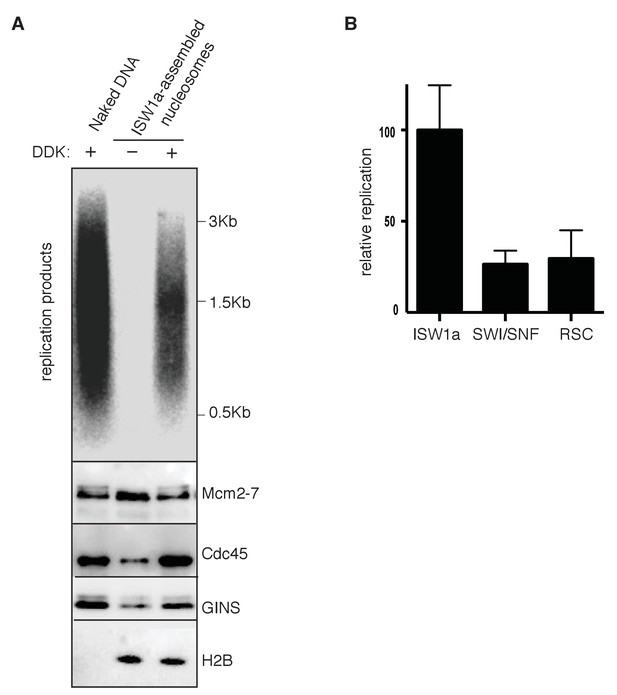
Reconstituted replication assay.
(A) Comparison of naked and nucleosomal DNA templates in the reconstituted replication assay. Comparison of replication of naked DNA and ISW1a templates with or without DDK using a fully reconstituted replication assay. Replication products (top panel) and Mcm2-7, Cdc45, GINS and H2B template association (lower panels) were assayed as in Figure 4D. (B) Comparison of nucleosomal DNA assembled with different CRE in the reconstituted replication assay. Quantification of the ISW1a, SWI/SNF and RSC templates replication products in the presence of DDK using reconstituted replication assay. Reconstituted replication assays performed as described in Figure 4D. Quantified as in Figure 3B and the material and method section. Error bars indicate standard deviation of three biological replicates (n = 3).
-
Figure 4—figure supplement 1—source data 1
Raw values used in the quantification of Figure 4B (n = 3).
- https://doi.org/10.7554/eLife.22512.018
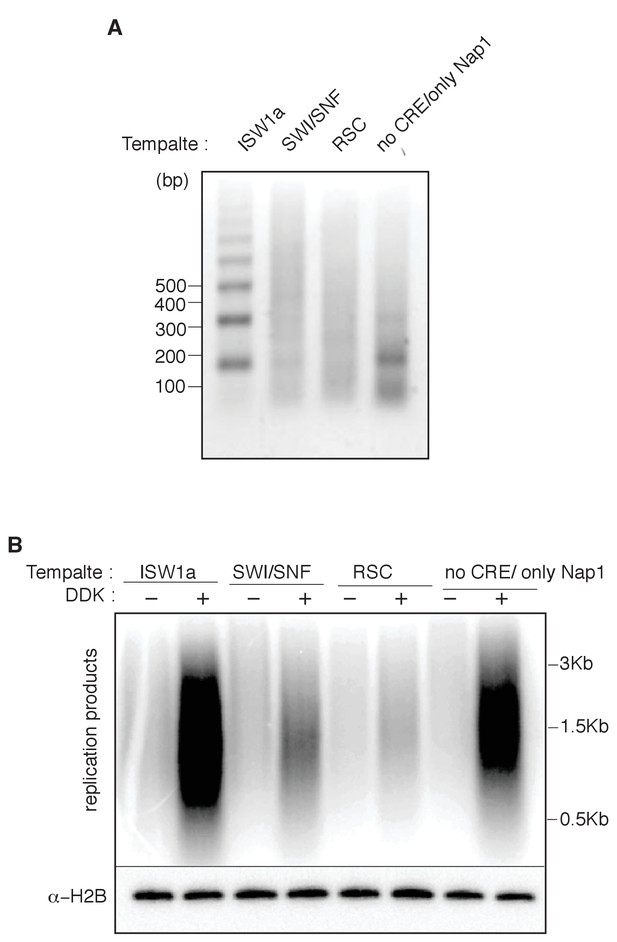
Nucleosomal template assembled without CRE are able to replicate.
(A) Nucleosomes were assembled with either ISW1a, SWI/SNF, RSC. or no CRE, with bead-coupled ARS1-containing linear DNA, yeast histone octamers and Nap1. Nucleosome assembly was assessed after partial MNase digestion as in Figure 2A. (B) Comparison of ISW1a, SWI/SNF, RSC or no CRE nucleosomal DNA templates in the reconstituted replication assay with or without DDK. H2B template association (lower panels) were assayed as in Figure 4D.
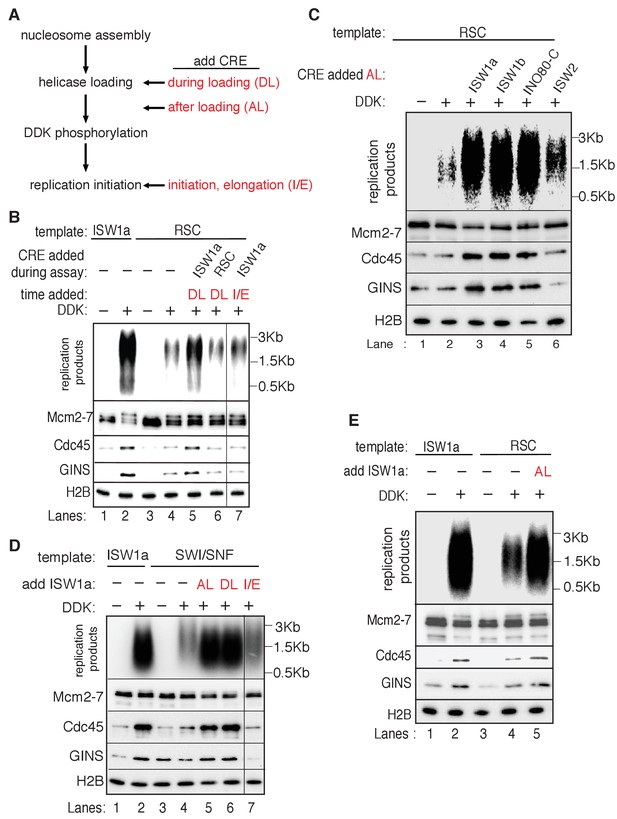
Rescue of SWI/SNF and RSC template replication initiation.
(A) Schematic of ISW1a addition at various steps during the replication assay. (B) Addition of ISW1a at the helicase-loading step rescues replication initiation from RSC templates. Reconstituted replication assays were performed on ISW1a and RSC templates with or without DDK. ISW1a or RSC was added to the templates during helicase loading and not deliberately removed (DL) or upon addition of the helicase activation and elongation proteins (I/E) as indicated. The lane that show I/E is from the same gel as the rest of the panel. Replication products (top panel) and Mcm2-7, Cdc45, GINS and H2B template association (lower panels) were assayed as in Figure 4D. (C) Specific CREs improve RSC-template replication. Reconstituted replication assays were performed with RSC templates with or without DDK. ISW1a, ISW1b, INO80-C or ISW2 was added to RSC templates after helicase loading (AL). Replication products (top) and Mcm2-7, Cdc45, GINS and H2B template association (lower panels) were assayed as in Figure 4D. (D) Addition of ISW1a after nucleosome assembly facilitates replication and CMG formation of SWI/SNF templates. Reconstituted replication assays were performed with ISW1a or SWI/SNF templates with or without DDK. ISW1a was added to the templates either during helicase loading (DL), after helicase loading (AL) or upon addition of the helicase activation and elongation proteins (I/E) as indicated. The lane that show I/E is from the same gel as the rest of the panel. Replication products (top) and Mcm2-7, Cdc45, GINS and H2B template association (lower panels) were assayed as in Figure 4D. (E) ISW1a addition after helicase loading (AL) to RSC templates, but removed before helicase activation improves replication of and CMG complex formation on RSC templates. Reconstituted replication reactions were performed with the indicated templates with or without DDK. ISW1a was added to the RSC templates upon completion of helicase loading (AL). Replication products (top) and Mcm2-7, Cdc45, GINS and H2B template-association (lower panels) were assayed as in Figure 4D.
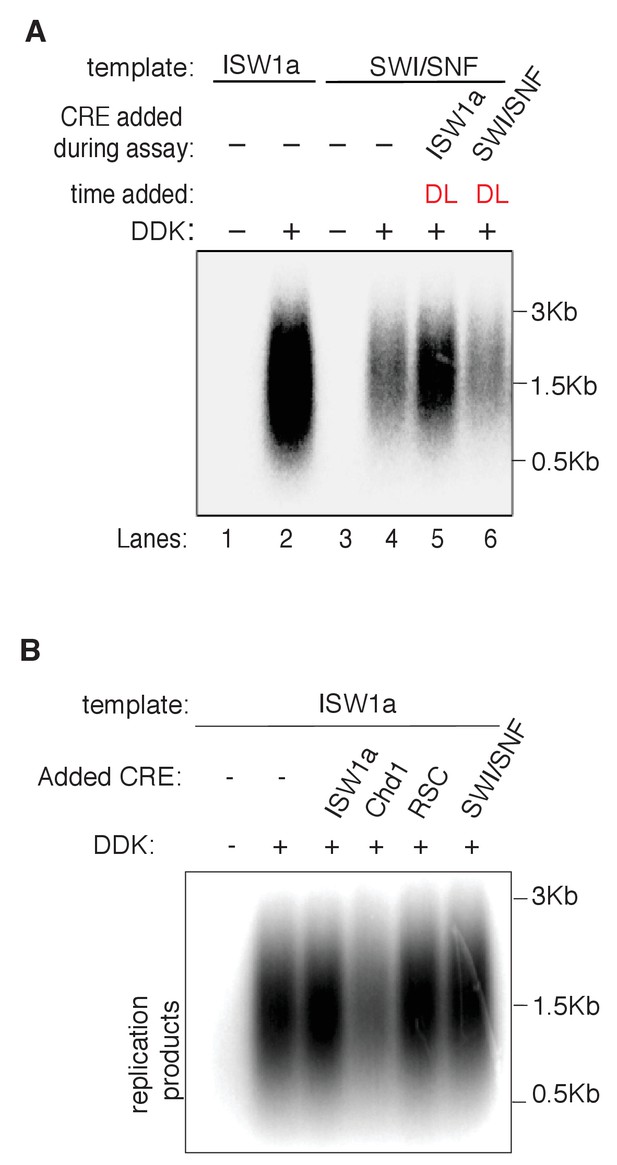
ISW1a rescues RSC and SWI/SNF templates prior to the initiation step.
(A) Addition of ISW1a to SWI/SNF templates, at helicase-loading step rescues SWI/SNF initiation defects. Reconstituted replication assays were performed on. ISW1a and SWI/SNF templates with or without DDK. ISW1a or SWI/SNF was added to the SWI/SNF templates during helicase loading (DL). Replication products were assayed as in Figure 3B. (B) Addition ISW1, Chd1, SWI/SNF or RSC to ISW1a templates, post at helicase-loading step. Reconstituted replication assays were performed on ISW1a templates with or without DDK. Replication products were assayed as in Figure 3B.
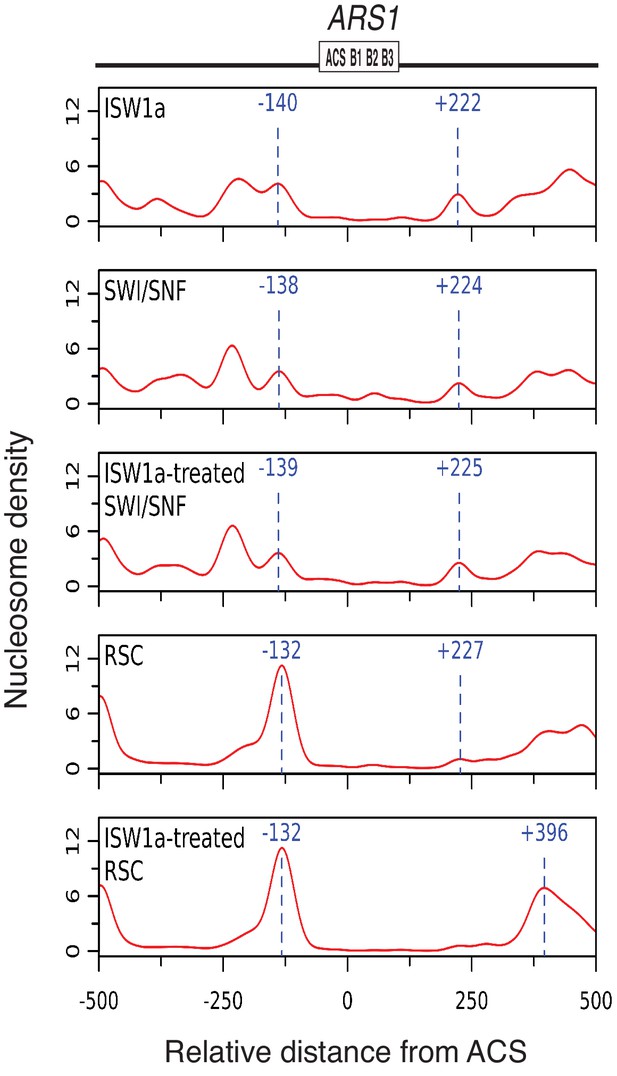
Origin proximal nucleosome positioning is not directly responsible for CMG formation defects in RSC and SWI/SNF templates.
Analysis of origin-proximal nucleosome positioning of ISW1a, SWI/SNF and RSC templates. Comparison of origin-proximal nucleosomes positioning by high-throughput MNase-Seq of ISW1a-, SWI/SNF- and RSC-templates similar to Figure 2C. RSC and SWI/SNF templates were remodeled with ISW1a (ISW1a-treated RSC or SWI/SNF) after they were assembled for indicated reactions. Zero on the X-axis indicates the first nucleotide of the ARS consensus sequence (ACS).
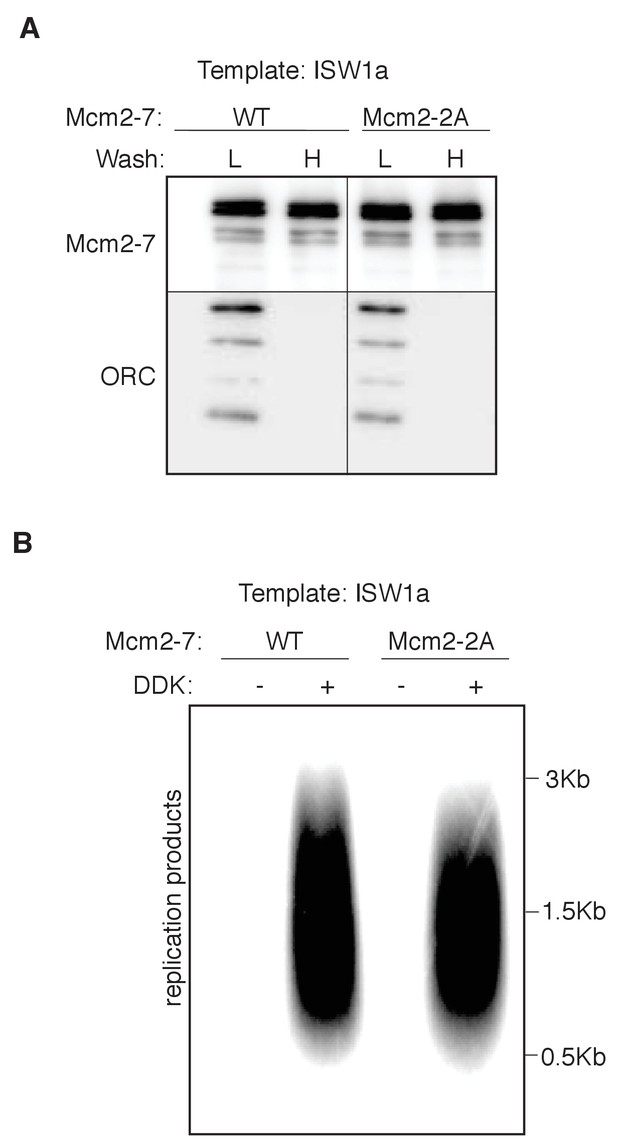
Mcm2 histone-binding motif is dispensable for nucleosomal DNA replication.
(A) Comparison of WT and mutant Mcm2 helicase loading on ISW1a-remodeled DNA templates. DNA templates were washed with high-salt (H) or low-salt (L) buffer after loading. Template-associated Mcm2-7 and ORC was detected by immunoblot. (B) WT and mutant Mcm2 helicase loading on ISW1aremodeled DNA templates were assayed for replication initiation as described for Figure 4B.
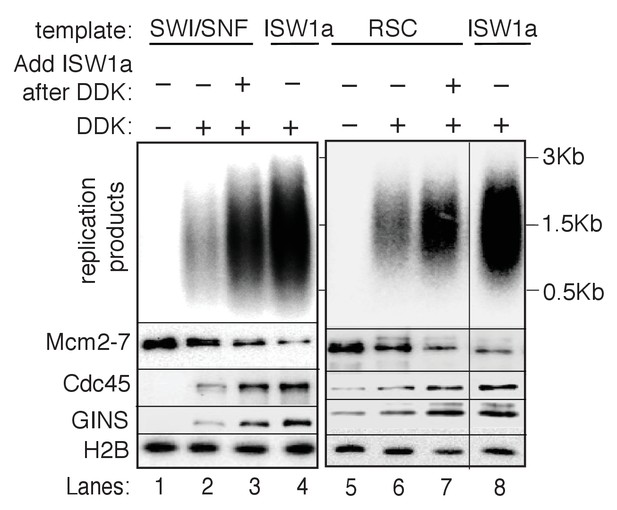
ISW1a rescues RSC and SWI/SNF templates after DDK step.
ISW1a addition to RSC and SWI/SNF templates after DDK phosphorylation step, partially restores RSC and SWI/SNF templates replication and CMG formation defects. Reconstituted replication assays were performed with the indicated templates in the presence and absence of DDK. ISW1a was added to the SWI/SNF and RSC templates upon completion of DDK-phosphorylation step and washed off before adding initiation factors. Replication products (top panel) and Mcm2-7, Cdc45, GINS and H2B template-association (lower panels) were assayed as in Figure 4D.
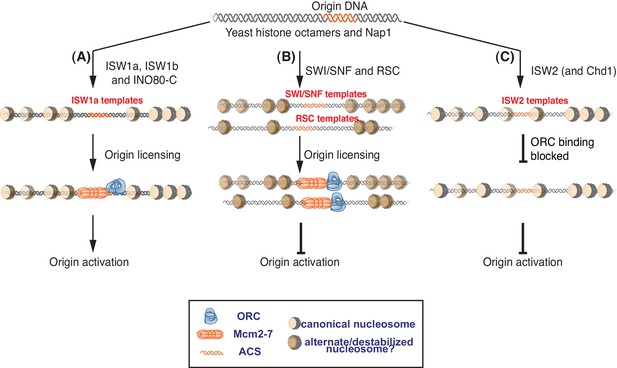
Nucleosomes remodeled by different CREs influence replication initiation differently.
Nucleosomes affect multiple steps of replication initiation using distinct mechanisms. Schematic of ATP-dependent nucleosome assembly with different CREs and their affect on replication initiation. Opacity of the nucleosome represents nucleosome density at each location. (A) Replication permissive nucleosomes are remodeled by ISW1a, ISW1b and INO80-C. These templates are competent for both origin licensing and origin activation. Nucleosome positioning is comparable in these templates. (B) SWI/SNF and RSC templates are origin-licensing competent but are inefficient for subsequent origin activation. We propose that the SWI/SNF and RSC templates have alternate/destabilized nucleosome structures indicated by their different color and that these nucleosomes are not conducive to origin activation. Although both reduce origin activation, SWI/SNF and RSC templates do not share similar nucleosome positioning. (C) ISW2 (and Chd1) templates have nucleosomes over the replication origin that reduce ORC DNA binding and, therefore, origin licensing.
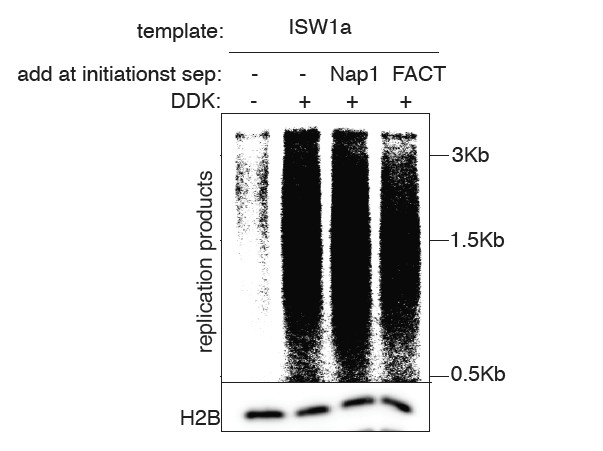
Additional files
-
Supplementary file 1
Yeast strains used in the study.
- https://doi.org/10.7554/eLife.22512.026
-
Supplementary file 2
Plasmids used in the study.
- https://doi.org/10.7554/eLife.22512.027





