Global analysis of gene expression reveals mRNA superinduction is required for the inducible immune response to a bacterial pathogen
Figures
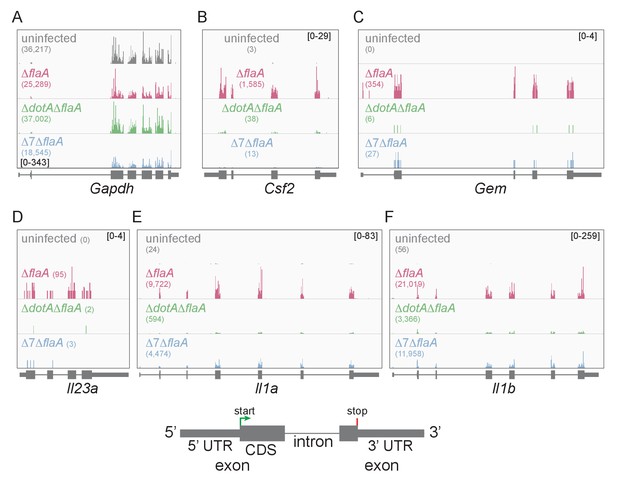
Mapping of ribosome profiling reads to the genomic sequence of specific L. pneumophila-induced genes of interest.
(A–F) Ribosome footprint reads were mapped to the genome and the number of footprints on the mRNAs for Gapdh (A), Csf2 (B), Gem (C), Il23a (D), Il1a (E), and Il1b (F) was visualized. Numbers in parentheses show the total read count of ribosome footprints found on the indicated transcript. Bracketed numbers represent read count data range. Gray, uninfected BMMs. Red, ΔflaA-infected BMMs. Green, ΔdotAΔflaA-infected BMMs. Blue, Δ7ΔflaA-infected BMMs.
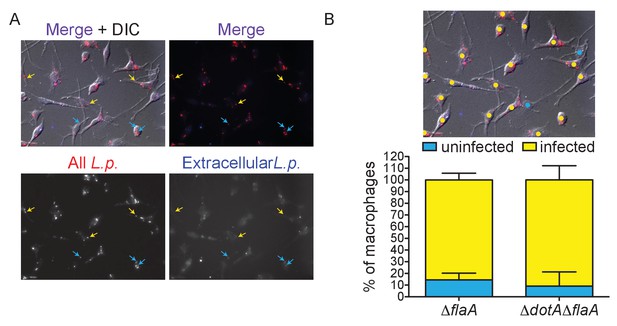
Quanification of L. pneumophila infectivity.
(A–B) ΔflaA or ΔdotAΔflaA L. pneumophila-infected BMMs were stained to mark extracellular (blue) and all bacteria (red). (A) Representative image of ΔflaA L. pneumophila-infected BMMs showing extracellular (blue and red stain, blue arrow) and intracellular (red stain only, yellow arrow) bacteria. (B) Individual BMMs (n = 1375) were analyzed for the presence of at least one intracellular ΔflaA or ΔdotAΔflaA L. pneumophila bacterium. Image is the same as in A with yellow circles marking infected cells and blue circles marking uninfected cells. The average combined infectivity in these conditions is ~90%. See supplemental methods for more details on counting.
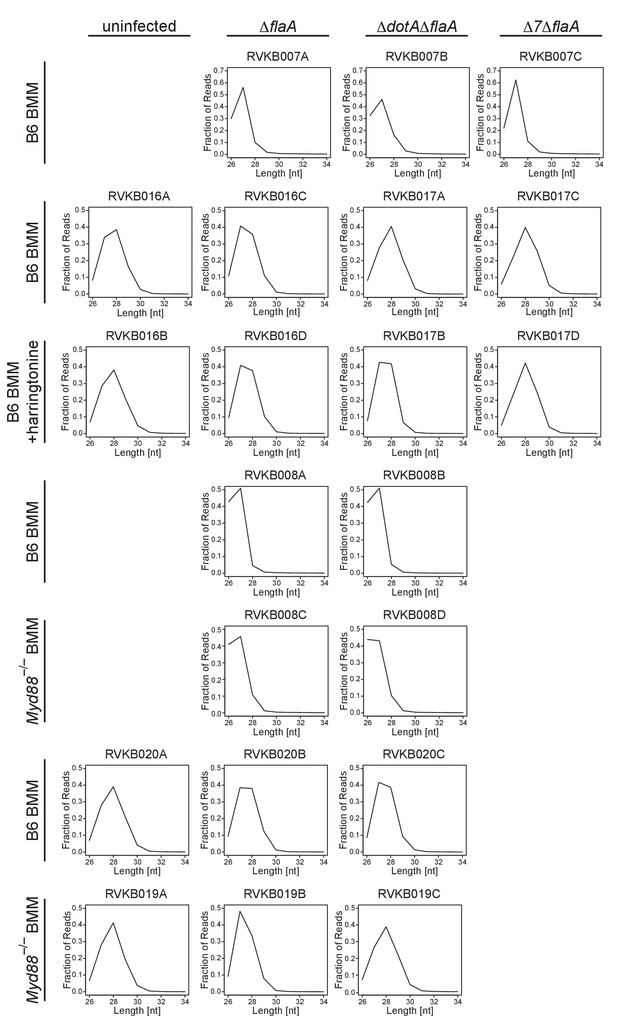
Ribosome profiling libraries show a strong bias in size distribution.
The fraction of total reads with a size of 26–34 nucleotides was plotted for each ribosome profiling library used in this study. These graphs clearly show that the ribosome profiling libraries used in this study have a strong bias for 27–28 nucleotide fragments, consistent with the size of the footprint of the ribosome. Columns indicate infection condition. Rows indicate BMM genotype and/or drug treatment.
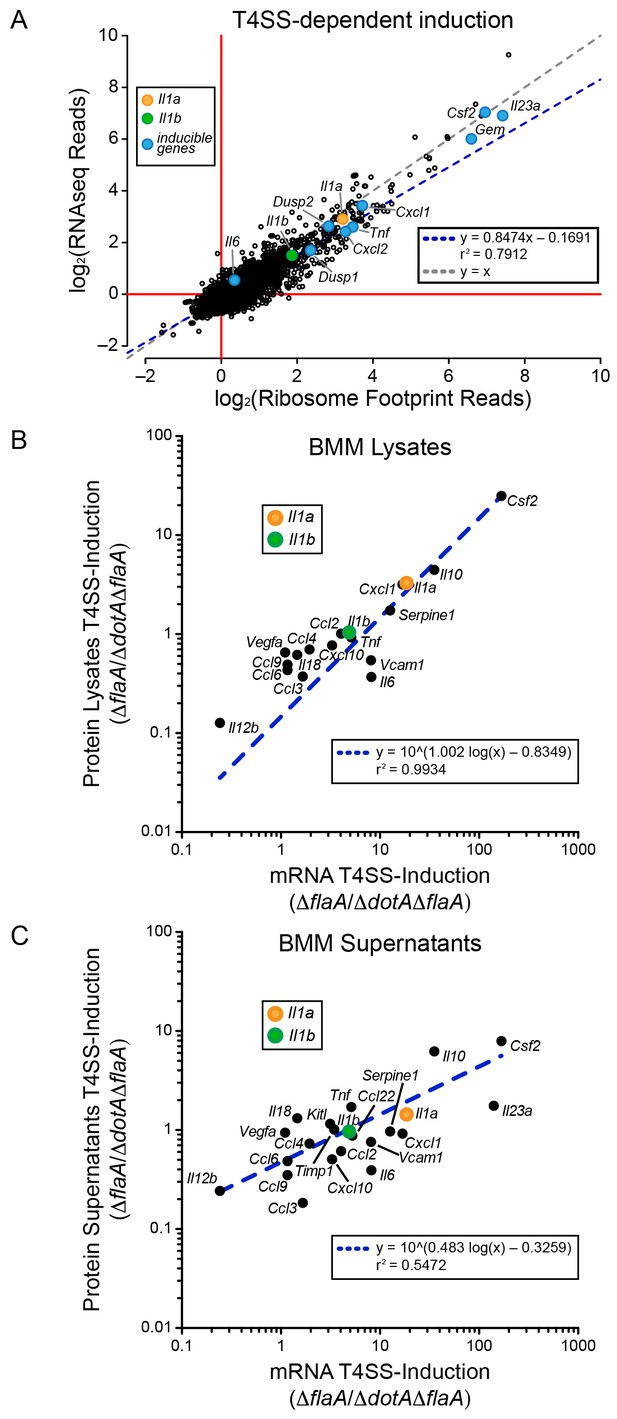
mRNA superinduction controls the T4SS-dependent induction of host gene expression in response to L. pneumophila.
(A) The ratio of ribosome footprint and RNAseq read counts for well-expressed transcripts (read count ≥100) in ΔflaA-infected versus ΔdotAΔflaA-infected B6 BMMs was calculated for each annotated transcript (open circles) in the dataset and plotted. (B–C) B6 BMMs were infected with ΔflaA or ΔdotAΔflaA L. pneumophila and at 6 hr post-infection proteins were measured in cell lysates (B) or supernatants (C) by bead array. The T4SS-induction (ΔflaA/ΔdotAΔflaA) of protein in supernatants (B) or lysates (C) and the T4SS-induction of mRNA (ΔflaA/ΔdotAΔflaA) was plotted. Proteins were normalized to total protein levels measured by BCA and RNAseq read counts was normalized to transcript length and the sum of their respective mitochondrial protein coding genes. Data are averaged from four (A) or two independent experiments (B–C). Orange circle, Il1a. Green circle, Il1b. Blue circle, subset of inducible genes. Grey dotted line, y = x. Blue dotted line, linear regression model. r2, coefficient of determination. See also Figure 2—source data 1.
-
Figure 2—source data 1
Source data from ribosome profiling, RNAseq, and bead array analysis used for Figure 2.
- https://doi.org/10.7554/eLife.22707.007
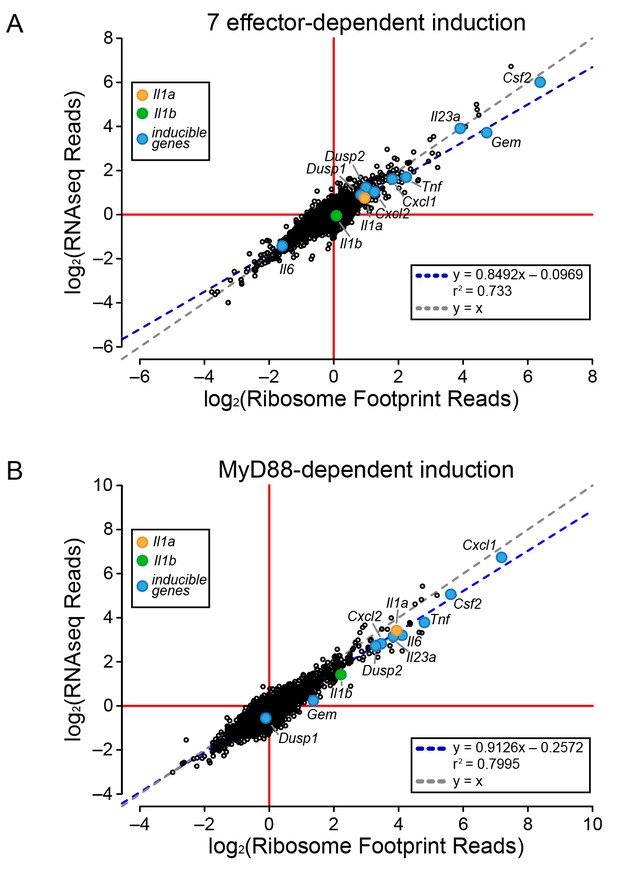
Global induction of mRNAs and ribosome footprints in response to L. pneumophila.
(A–B) Ribosome footprint and RNAseq read counts were sorted for well-expressed transcripts (read count ≥100) and normalized to the sum of their respective mitochondrial protein coding genes. The ratio of ribosome footprint and RNAseq read counts in (A) ΔflaA-infected and Δ7ΔflaA-infected B6 BMMs or (B) B6 or Myd88–/– BMMs infected with ΔflaA L. pneumophila was calculated for each annotated transcript (open circles) in the dataset and plotted. Data are averaged from two independent experiments. Orange circle, Il1a. Green circle, Il1b. Blue circle, subset of inducible genes. Grey dotted line, y = x. Blue dotted line, linear regression model. r2, coefficient of determination. See also Figure 3—source data 1.
-
Figure 3—source data 1
Source data from ribosome profiling and RNAseq analysis used for Figure 3.
- https://doi.org/10.7554/eLife.22707.009
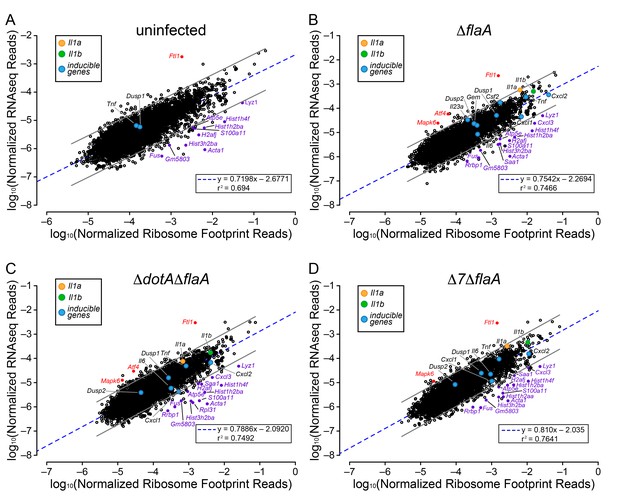
Ribosome occupancy does not explain the inducible innate immune response to L. pneumophila.
(A–D) Ribosome footprint and RNAseq read counts were sorted for well-expressed transcripts (read counts ≥ 100) and normalized to CDS length and the sum of their respective mitochondrial protein coding genes. The normalized read counts for ribosome footprints and RNAseq for all well-expressed annotated transcripts were plotted for (A) uninfected, (B) ΔflaA, (C) ΔdotAΔflaA, or (D) Δ7ΔflaA L. pneumophila-infected B6 BMMs. Red dots represent transcripts with low translation efficiency. Purple dots represent a number of transcripts common to all conditions that appear to have significantly higher ribosome occupancy. Data are averaged from three (A), four (B–C), or two independent experiments (D). Orange circle, Il1a. Green circle, Il1b. Blue circles, subset of inducible transcripts. Blue dotted line, linear regression model. Grey lines, 99% prediction interval. r2, coefficient of determination. See also Table 1 and Figure 4—source data 1.
-
Figure 4—source data 1
Source data from ribosome profiling and RNAseq analysis used for Figure 4.
- https://doi.org/10.7554/eLife.22707.011
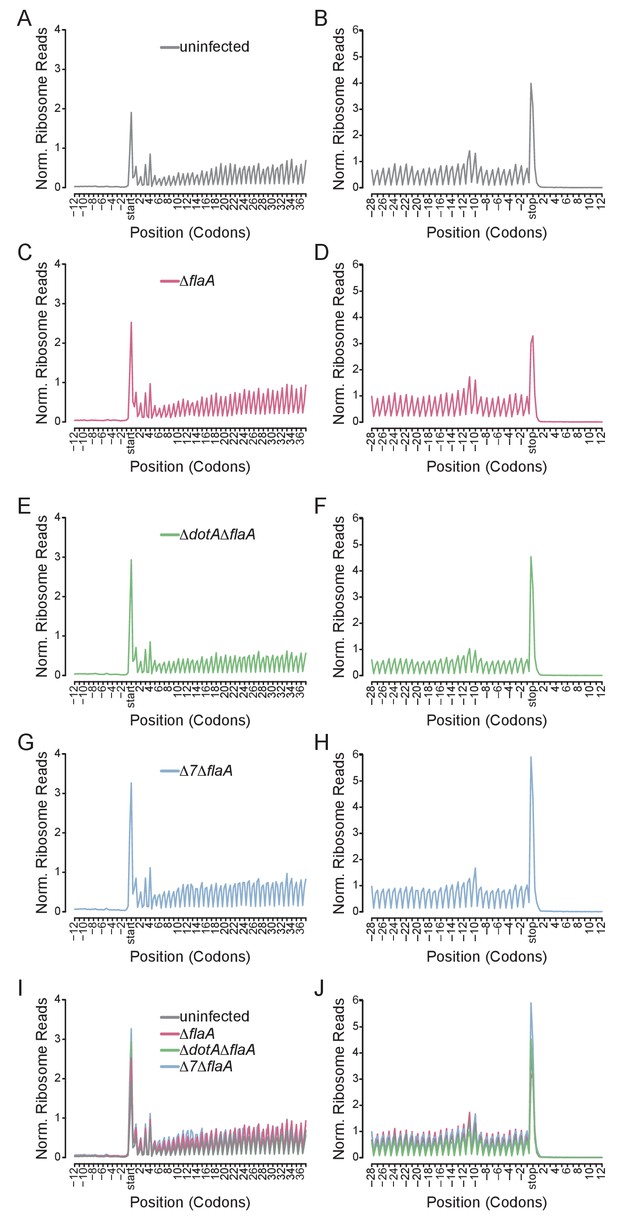
Metagene profiles of L. pneumophila infected BMMs.
(A–J) Metagene profiles of uninfected (A–B), ΔflaA (C–D), ΔdotAΔflaA (E–F), Δ7ΔflaA (G–H) L. pneumophila-infected B6 BMMs and a merge (I–J). Metagene profiles are depicted relative to the translation start (A, C, E, G, I) and stop site (B, D, F, H, J). Metagene analyses show peaks at every three nucleotides, corresponding to the codon-to-codon shifts of the ribosome. Data are representative of two independent experiments (A–J). Black line, uninfected. Red line, ΔflaA-infected. Green line, ΔdotAΔflaA-infected. Blue line, Δ7ΔflaA-infected.
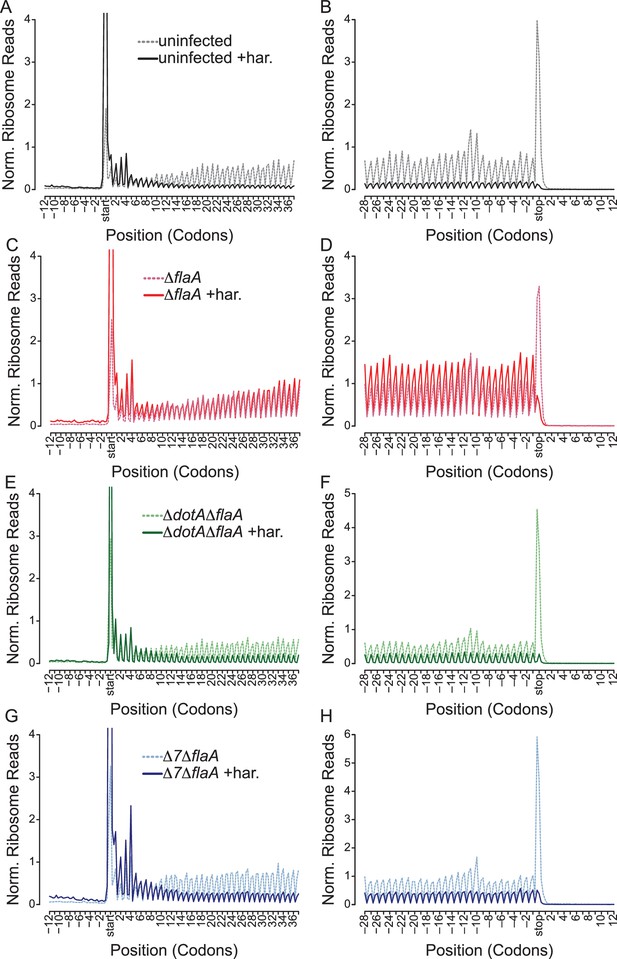
L. pneumophila-induced block of host protein synthesis occurs at the level of translation initiation and elongation.
(A–H) Metagene profiles of B6 BMMs uninfected (A–B) or infected with ΔflaA (C–D), ΔdotAΔflaA (E–F), or Δ7ΔflaA (G–H) L. pneumophila in the presence (solid line) or absence (dashed line) of the drug harringtonine to block translation initiation. Metagene profiles are depicted relative to the translation start (A, C, E, G) and stop site (B, D, F, H). Data are representative of two independent experiments (A–H). Solid line, no drug treatment. Dashed line, harringtonine treatment. Black line, uninfected. Red line, ΔflaA-infected. Green line, ΔdotAΔflaA-infected. Blue line, Δ7ΔflaA-infected.
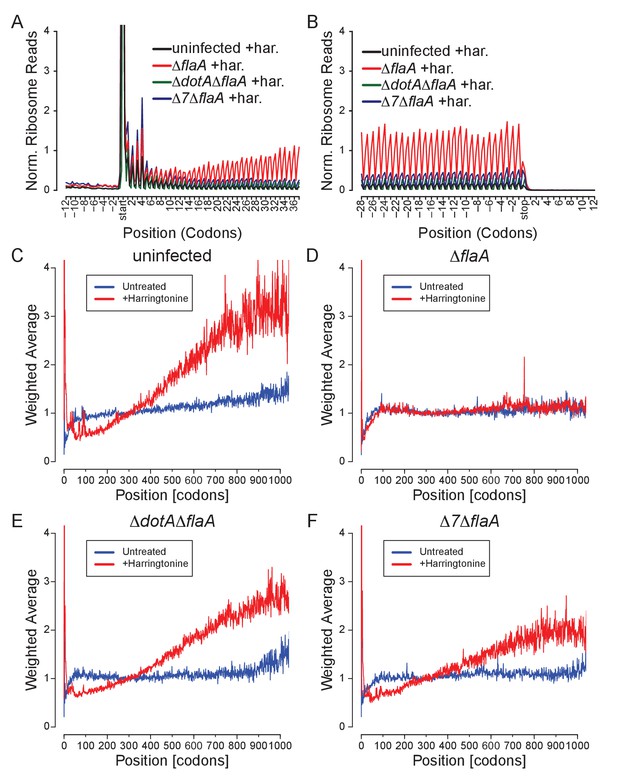
L. pneumophila-induced block in host protein synthesis can occur at the level of translation elongation and initiation.
(A–B) Metagene profile plot around the translation start (A) or stop (B) site of all harringtonine-treated conditions normalized to mitochondrial read counts of each condition. (C–F) Global weighted averages across transcripts were calculated for BMMs left uninfected (C) or infected with ΔflaA (D), ΔdotAΔflaA (E), or Δ7ΔflaA (F) L. pneumophila. Weighted averages were generated by scaling each transcript’s ribosome occupancy profile according to the average density from codon 250 to codon 349 and then averaging across the entire condition. Transcripts with very low density in the 250–349 codon region (or shorter than 349 codons) are excluded from averaging. If the weighted average is less than 1, this shows that this region has reduced ribosome footprints, while if the weighted average is greater than one this shows that there are more ribosome footprints in this region. Following a brief pulse of cells with harringtonine (red line) there is a change in the distribution of ribosomes from the 5ʹ end of the mRNA to the 3ʹ end of the mRNA (i.e., as the ribosomes continue to move 5ʹ to 3ʹ; C–F) as compared to untreated cells (blue line). This can be seen by an increase in the weighted average at the 3ʹ end of mRNAs (C, E, F) but the lack of this change shows a block in translation elongation (D). We also expect an accumulation of ribosomes at the ATG following harringtonine treatment, which is also seen as a peak in the weighted average at the start site (C–F). Data are representative of two independent experiments (A–F).
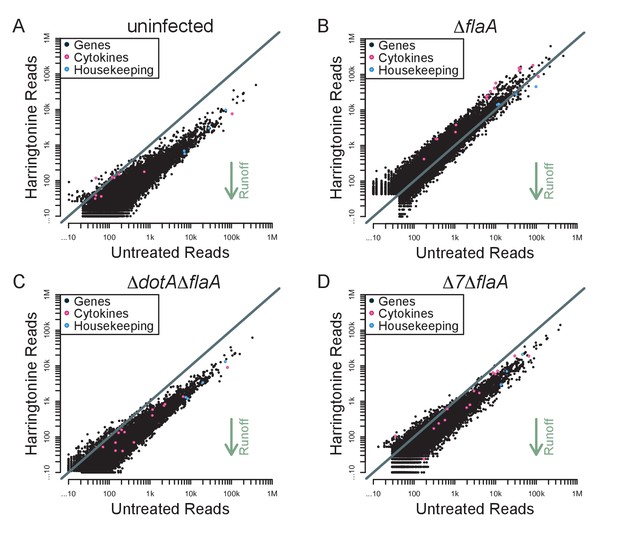
Immune-related genes do not have increased translation rates in response to infection with L. pneumophila.
(A–D) Read counts from paired samples treated with harringtonine or left untreated were plotted for uninfected (A), ΔflaA (B), ΔdotAΔflaA (C), or Δ7ΔflaA-infected (D) BMMs showing where cytokine-related transcripts (pink circles; Csf1, Csf2, Cxcl1, Cxcl2, Dusp1, Dusp2, Ifnb1, Il10, Il12b, Il1a, Il1b, Il23a, Il6, Lyz1, and Tnf) and housekeeping transcripts (blue circles; Gapdh, Rpl31, Rps17, and Tuba1a) fall among all transcripts (black circles). Grey line, y=x. Data shown are representative of two independent experiments. See Figure 7—source data 1 for individual housekeeping and cytokine-related transcripts. Supporting Information Captions.
-
Figure 7—source data 1
Source data from ribosome profiling analysis used for Figure 7.
- https://doi.org/10.7554/eLife.22707.017
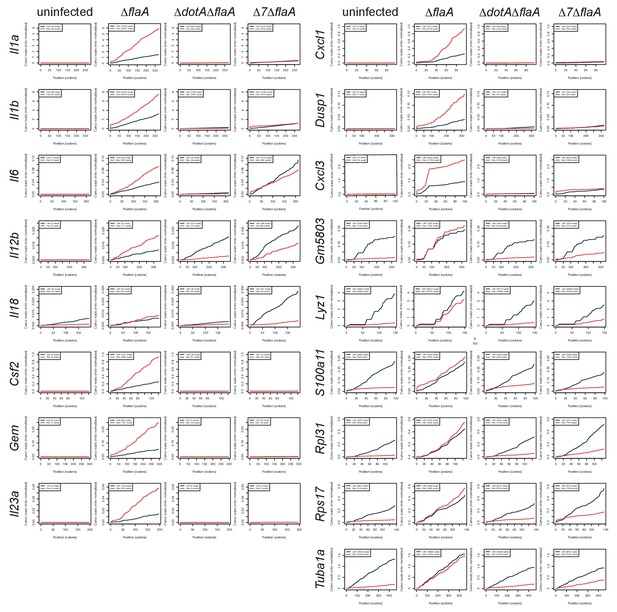
Individual mRNAs do not show evidence of preferential translation.
Cumulative read counts across the length of individual transcripts were calculated and normalized to the sum of ribosome footprints of mitochondrial transcripts in each respective library. Red = BMMs treated with harringtonine. Black = BMMs untreated. Numbers in parentheses = total read counts. Rows = individual transcript. Columns = infection condition.
Tables
Transcripts with ribosome occupancy eight times greater than the condition average. Bolded, transcripts found in all conditions. Orange, transcripts found in three conditions. Purple, transcripts found in two conditions. Data are averaged from two independent experiments.
| Uninfected | ΔflaA | ΔdotAΔflaA | Δ7ΔflaA | ||||
|---|---|---|---|---|---|---|---|
| Gene | Riobosome occupancy | Gene | Riobosome occupancy | Gene | Riobosome occupancy | Gene | Riobosome occupancy |
| Acta1 | 7105.62 | Acta1 | 3137.40 | Acta1 | 2489.70 | Acta1 | 1633.44 |
| H2-Q7 | 3130.45 | Hist1h4f | 1177.90 | S100a11 | 1115.36 | Hist1h4f | 943.87 |
| Hist1h4f | 2195.70 | S100a11 | 870.51 | Hist1h4f | 1069.93 | Rpl31 | 893.93 |
| Hist3h2ba | 1715.98 | Hist1h2aa | 844.87 | Rpl31 | 873.61 | Hist1h2aa | 822.96 |
| H2afj | 1524.09 | Hist3h2bb-ps | 699.92 | Hist1h2ba | 707.62 | Hist3h2ba | 625.90 |
| Hist3h2bb-ps | 1470.88 | Hist1h2ba | 692.19 | Hist3h2ba | 670.32 | S100a11 | 565.91 |
| Lyz1 | 1260.16 | H2afj | 686.47 | Lyz1 | 533.22 | Fus | 532.18 |
| Hist1h2ba | 1174.16 | Cxcl3 | 675.23 | Hist3h2bb-ps | 524.22 | Hist1h2ba | 519.60 |
| Cd52 | 1170.22 | H2-T24 | 557.88 | Fus | 405.23 | Lyz1 | 498.70 |
| Fus | 1102.13 | Lyz1 | 551.76 | H2-T24 | 374.06 | H2-Q7 | 371.10 |
| H2-Q4 | 1022.17 | Hist3h2ba | 509.95 | Gm5803 | 345.78 | Gm5803 | 368.22 |
| Rpl38 | 1004.74 | Fus | 480.25 | H2-Q7 | 337.36 | H2afj | 356.99 |
| Hist2h2ab | 796.60 | Saa1 | 475.25 | Hist1h4i | 302.55 | Hist1h4i | 348.34 |
| H2-Q6 | 752.77 | Gm5803 | 436.48 | Cxcl3 | 281.86 | Hist1h4k | 318.08 |
| S100a11 | 717.99 | Hist1h4i | 333.72 | Saa1 | 265.08 | Rrbp1 | 306.19 |
| Gm5803 | 692.32 | Atp5e | 315.00 | Hist1h4n | 244.74 | Hist1h4j | 301.73 |
| Tmsb10 | 679.27 | Rrbp1 | 308.68 | Rrbp1 | 225.85 | Hist1h4a | 298.67 |
| H2-Q10 | 674.79 | Mt1 | 308.36 | Hist1h4j | 218.84 | Hist1h4h | 295.99 |
| Rpl36 | 672.43 | Hist1h4j | 304.87 | Hist1h4k | 217.89 | Hist1h4b | 288.98 |
| Mt1 | 672.29 | Hist1h4k | 303.19 | Atp5e | 217.08 | Hist1h4n | 272.04 |
| Hist2h2bb | 650.90 | Hist1h4h | 293.11 | H2-Q4 | 215.94 | BC094916 | 259.08 |
| H2-Q7 | 629.56 | Hist1h4a | 292.51 | Hist1h4h | 215.51 | Hist1h4c | 255.20 |
| H2-Q7 | 618.80 | Hist1h4b | 280.60 | H2afj | 206.14 | Cxcl3 | 252.49 |
| Atp5e | 606.27 | Gm11127 | 272.59 | Hist1h4a | 205.64 | Atp5e | 241.23 |
| H2-T24 | 601.41 | Hist1h4n | 265.41 | Hist1h4b | 197.15 | Saa1 | 220.10 |
| Rpl37 | 584.88 | Fkbp1a | 264.22 | Hist2h2bb | 191.90 | Myl12b | 210.72 |
| H2-T10 | 545.54 | Hist1h4c | Hist1h4c | Hist1h4c | 187.03 | Gm7030 | 206.93 |
| Hist1h4i | 529.15 | Gm7030 | 253.47 | Mt1 | 185.28 | ||
| Gm11127 | 512.74 | Myl12b | 247.71 | Cd52 | 184.14 | ||
| Uqcrq | 511.93 | Rps17 | 234.41 | Gm11127 | 183.08 | ||
| Emp1 | 494.39 | Cd52 | 231.77 | Hist1h2bj | 182.74 | ||
| Hist1h2bf | 484.53 | Sh3bgrl | 181.81 | ||||
| Gm7030 | 481.28 | ||||||
| Npc2 | 479.93 | ||||||
| Hist1h2bj | 478.33 | ||||||
| Usmg5 | 477.21 | ||||||
| Hmga2 | 468.10 | ||||||





