Mice deficient of Myc super-enhancer region reveal differential control mechanism between normal and pathological growth
Figures
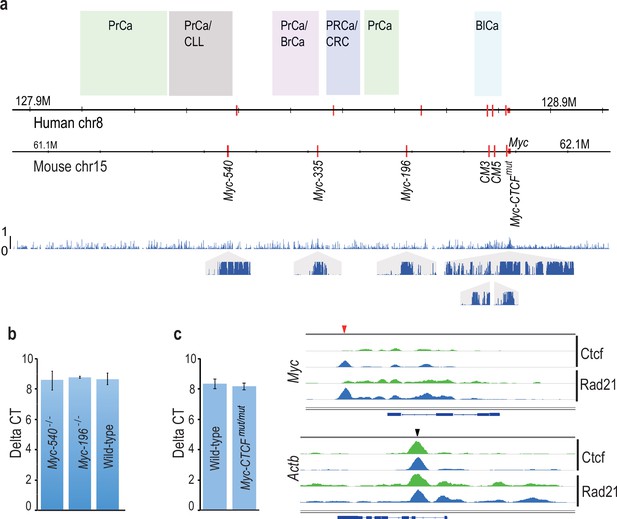
Cancer susceptibility region upstream of Myc contains several conserved enhancer elements that are dispensable for normal mouse development and MYC expression.
(a) Comparison of Myc locus between human and mouse. The susceptibility regions for prostate cancer (PrCa), chronic lymphocytic leukemia (CLL), breast cancer (BrCa), colorectal cancer (CRC) and bladder cancer (BlCa) are marked. Red vertical lines mark the location of the Tcf7l2-binding CRC Myc enhancers in the two species. The lower panel shows the regional conservation probability predicted by PhastCons (hg19 assembly, UCSC) with non-overlapping sliding windows for the whole region and each enhancer locus with a size of 500 bp and 10 bp, respectively. (b) Deletion of Myc-196 and Myc-540 enhancer elements does not affect Myc expression in the colon as determined by qPCR analysis (Myc-196−/− n = 2, Myc-540−/− n = 3 and wild-type n = 5). See Figure 1—source data 1 for details. (c) Mutation of the Myc-CTCF site causes loss of CTCF and Rad21 binding at the Myc locus (top panel). Binding of CTCF and Rad21 at a control Actb locus is not affected. Red and black arrowheads denote binding sites at Myc and Actb loci, respectively; green: Myc-CTCFmut/mut, blue: wild-type. The gene body for Myc and Actb is shown below the respective panels. The qPCR analysis reveals that despite loss of CTCF/cohesin binding, the expression of Myc mRNA is not altered in the colon (for qPCR, Myc-CTCFmut/mut n = 4, wild-type n = 3). See Figure 1—source data 1 for details. Error bars denote one standard deviation.
-
Figure 1—source data 1
Myc transcript levels in wild-type and mutant mice in Figure 1b-c.
- https://doi.org/10.7554/eLife.23382.004
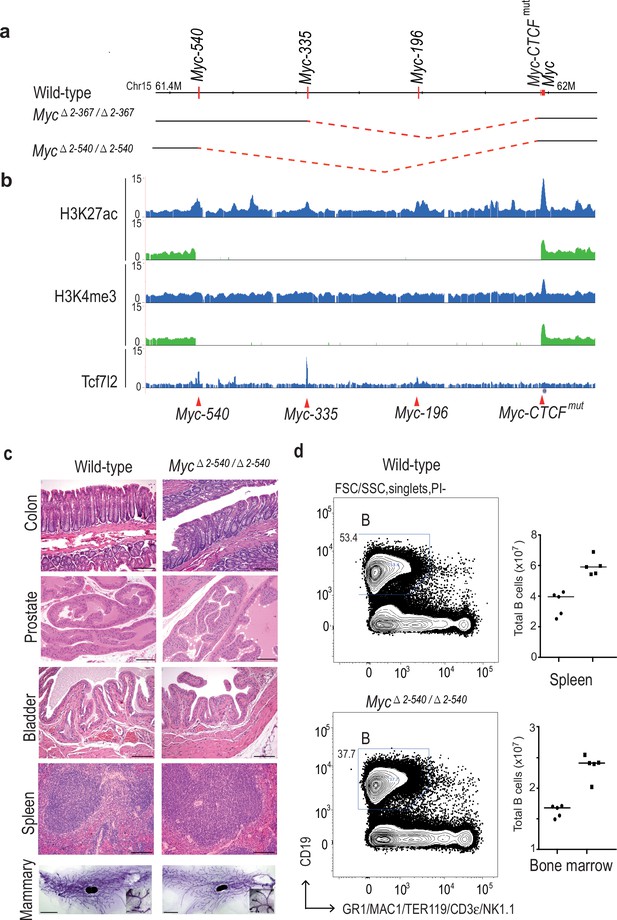
Deletion of the 8q24 super-enhancer region is well tolerated during normal development and homeostasis.
(a) Schematic representation of the 365 kb and 538 kb deletions. (b) Myc△2–540/△2–540 deletion removes a region containing several active enhancer elements upstream of Myc as shown by ChIP-seq analysis of histone H3 lysine 27 acetylation (H3K27ac) and lysine four trimethylation (H3K4me3). The deletion also removes several Tcf7l2 ChIP-seq peaks. Signal from Myc△2–540/△2–540 and wild-type mice are shown in green and blue, respectively. Red arrowheads and horizontal lines mark the different enhancer positions. (c) Haematoxylin/ Eosin stained sections of spleen, bladder, prostate, colon (Bar = 100 µm) and Carmine Alum stained whole mounts of mammary glands, Bars = 3 mm, 100 µm (inset) showing normal development and homeostasis of different organs in Myc△2–540/△2–540 mice. (d) Myc△2–540/△2–540 mice have a reduced number of B-cells compared to the wild-types. Left panel: FACS plots of a representative Myc△2–540/△2–540 and wild-type mouse spleen showing B-cell (B) population. Right panel: Scatter dot plot of total number of B cells in the spleen and bone marrow of wild-type (squares), n = 5 and Myc△2–540/△2–540 (filled circles), n = 5. Each point represents individual mouse. Line represents the median. See Figure 2—source data 1 for details. The number of CD4+ and CD8+ T-cells is not affected by the deletion (see Figure 2—figure supplement 1 and appendix 1).
-
Figure 2—source data 1
B cell numbers in the wild-type and MycΔ2-540/Δ2-540 mice in Figure 2d.
- https://doi.org/10.7554/eLife.23382.006
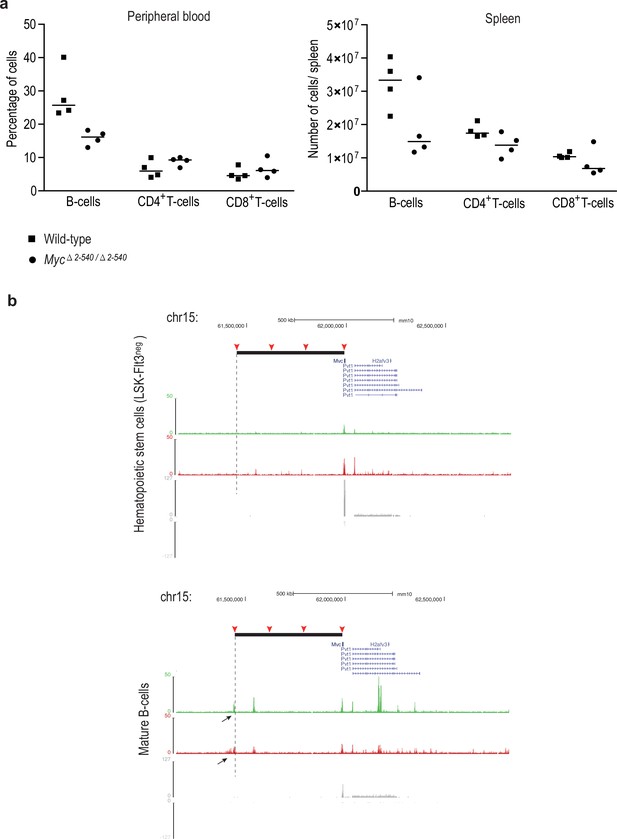
The loss of the Myc super-enhancer region results in a decrease in the number of B-cells, but no major defects in hematopoiesis.
(a) Super-enhancer region deletion results in a lower number of total cells in the spleen (average number in wild-type = 150.1×106 and Myc△2–540/△2–540 = 117.2×106) which manifests as a specific decrease of B-cell numbers both in the spleen and peripheral blood without an affect on the CD4+ and CD8+ T-cell population. Line marks the median. See Figure 2—figure supplement 1—source data 1 for details. (b) ChIP-seq analysis of histone marks H3K4me2 (red), H3K27ac (green) and RNA-seq (grey: signals from plus and minus strand are shown) shows the presence of B-cell specific enhancer both 3´and 5´ side of the Myc ORF. One of these enhancers (black arrow, mature B-cells panel) is located at the 5´boundary of the deleted region (black solid bar). The 2–540 deletion will bring this element very close to the Myc TSS, potentially explaining the increased Myc expression in the spleen. The different enhancer regions used in this study are marked by red arrowhead and the 5´ boundary of the super-enhancer region is marked by dashed line. The increase in MYC levels in the spleen and decrease in B-cell number could potentially be explained by a direct effect of MYC in inducing apoptosis of B-cells (Hoffman and Liebermann, 2008). However, further studies are necessary for dissection of the role of specific Myc enhancers during hematopoiesis.
-
Figure 2—figure supplement 1—source data 1
B and T-cell populations in the wild-type and MycΔ2-540/Δ2-540 mice in Figure 2—figure supplement 1a.
- https://doi.org/10.7554/eLife.23382.008
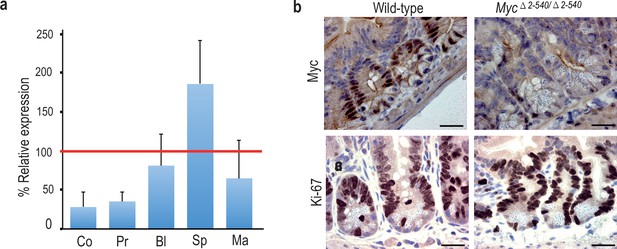
Tissue-specific effect of Myc△2–540/△2–540 deletion on MYC expression.
(a) qPCR data showing the percentage of Myc expression in Myc△2–540/△2–540 relative to the wild-type in colon (Co) n = 4, prostate (Pr) n = 2, bladder (Bl) n = 5, spleen (Sp) n = 4 and mammary gland (Ma) n = 3. Red line marks the expression level (100%) in wild-type mice. Error bars indicate one standard deviation. See Figure 3—source data 1 for details. (b) Immunohistochemistry shows reduced expression of MYC (n = 3 for each genotype) protein in intestinal crypts of Myc△2–540/△2–540 mice without any significant effect on proliferation as indicated by Ki-67 (n = 2 for each genotype) immunostaining, Bar = 10 µm. Brown: IHC staining, Blue: Haematoxylin staining.
-
Figure 3—source data 1
Myc transcript levels in MycΔ2-540/Δ2-540 mice relative to the wild-types in Figure 3a.
- https://doi.org/10.7554/eLife.23382.010
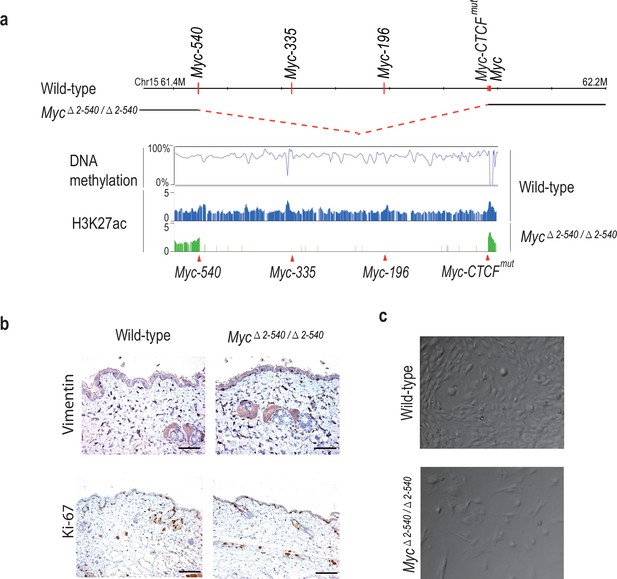
Myc△2–540/△2–540 deletion results in a proliferation defect of adult skin fibroblasts cultured in vitro.
(a) The super-enhancer region deleted in the Myc△2–540/△2–540 has under methylated DNA as determined through bisulfite sequencing of the wild-type fibroblasts grown in culture. H3K27ac ChIP-seq shows the presence of active enhancer marks within this region in the wild-type fibroblasts whereas the Myc△2–540/△2–540 fibroblasts show a complete absence of the super-enhancer region. The Myc super-enhancer region is also active in human fibroblasts (see Figure 4—figure supplement 1). (b) Normal morphology and proliferation of resident fibroblasts in the mouse skin as determined by Vimentin and Ki-67 IHC staining respectively in both the wild-type (n = 3) and Myc△2–540/△2–540 mice (n = 3), Bar = 50 µm. Brown: IHC staining, Blue: Haematoxylin staining (c) Representative phase contrast images of wild-type and Myc△2–540/△2–540 primary fibroblasts showing growth defect of Myc△2–540/△2–540 fibroblasts in culture.
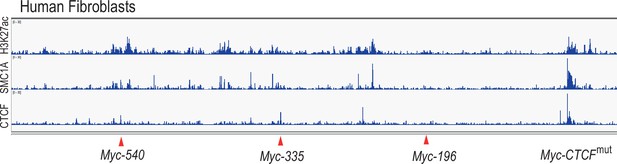
The Myc super-enhancer region is also active in human fibroblasts.
ChIP-seq analysis of H3 lysine 27 acetylation (H3K27ac), cohesion (SMC1A) and CTCF binding shows several active enhancer marks in the super-enhancer region upstream of the MYC gene in human fibroblasts. Red arrows mark the positions of the conserved enhancer elements used in this study.
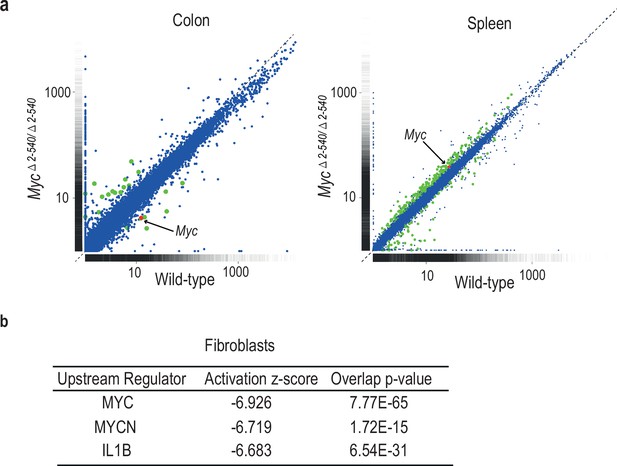
Differential effect of Myc△2–540/△2–540 deletion on MYC target gene expression.
(a) Scatter plot comparing the average Fragments per kilobase of exons per million fragments mapped (FPKM) values of gene transcripts in colon and spleen of wild-type (n = 4) and Myc△2–540/△2–540 (n = 4) mice. Genes showing significant (q < 0.05) differential expression are marked in red (Myc) or green (other genes). For median FPKM values of gene transcripts see Figure 5—figure supplement 1 (b) Upstream regulator analysis of RNA-seq data shows that the highest ranked potential regulator affected in the slow growing Myc△2–540/△2–540 fibroblasts is MYC. The activation z-scores are to infer the activation states of predicted upstream regulators. The overlap p-values were calculated from all the regulator-targeted differential expression genes using Fisher’s Exact Test. Two independent Myc△2–540/△2–540 fibroblasts lines were analysed to confirm the downregulation of Myc. Ingenuity pathway analysis performed on one of these is shown.
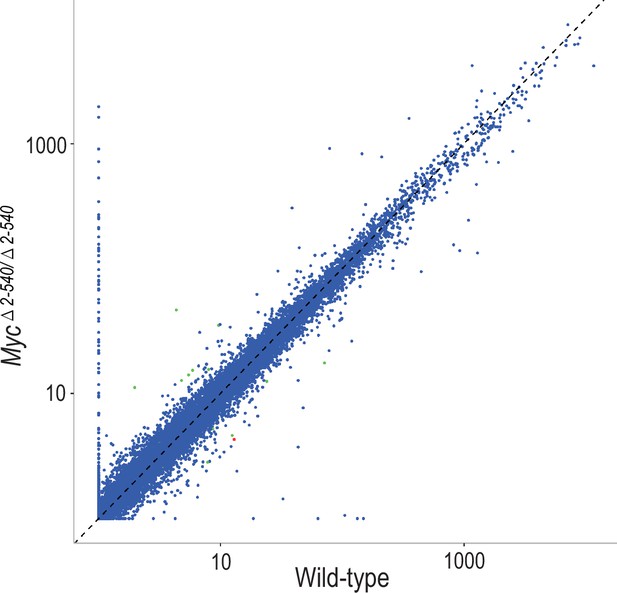
Scatter plot comparing the median of FPKM values of gene transcripts in colon of wild-type (n = 4) and Myc△2–540/△2–540 (n = 4) mice.
Genes showing significant (q < 0.05) differential expression are marked in red (Myc) or green (other genes). The plot was generated using ggplot2 (version 2.2.1, RRID:SCR_014601). The median FPKM values of gene transcripts were plotted for the colon data since one of the wild-type samples for unknown reason had higher amounts of ribosomal structural protein transcripts.
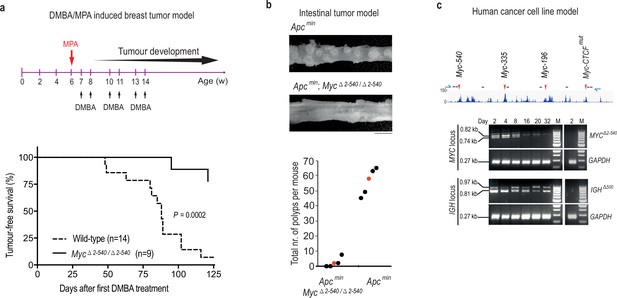
Myc −2 to −540 kb genomic region is required for the growth of cancers in vivo and cancer cells in vitro.
(a) Tumor-free survival plots showing resistance of Myc△2–540/△2–540 mice to development of DMBA/MPA induced mammary tumors. p-value=0.0002 (Mantel-Cox Log-rank test). See Figure 6—source data 1 for details. (b) The Myc −2 to −540 kb deletion results in fewer polyps than the Myc-335 deletion alone. p-value=0.00019 (Students T-test, 2-tailed). Apcmin mice were of 4 months of age (n = 5) and Apcmin; Myc△2–540/△2–540 mice were 6 months old (n = 5) at the time of analysis. Filled circles correspond to individual mice and red color denotes the median. See Figure 6—source data 1 for details. Bar equals 5 mm. (c) Crispr-Cas9 mediated deletion of region corresponding to Myc△2–540/△2–540 in human GP5d colon cancer cells, results in a loss of the edited cells over time. Top panel shows the active enhancer elements in GP5d cells within this region as determined by ChIP-seq analysis of histone H3 lysine 27 acetylation (H3K27ac). The sites of sgRNAs (black lines) and genotyping primers (blue arrows) used are indicated (not to scale). Red arrows mark the enhancer regions used in this study. Bottom panel shows the PCR-genotyping of the MYC locus and the control IGH locus showing the specific loss of the cells with the edited MYC locus over time. GAPDH was used as internal control. The right panel in each set shows absence of any deletion in the non-transfected cells (day 2). 100 bp ladder DNA molecular weight marker is shown (M).
-
Figure 6—source data 1
Survival time and intestinal polyp numbers for mice in Figure 6a-b.
- https://doi.org/10.7554/eLife.23382.016
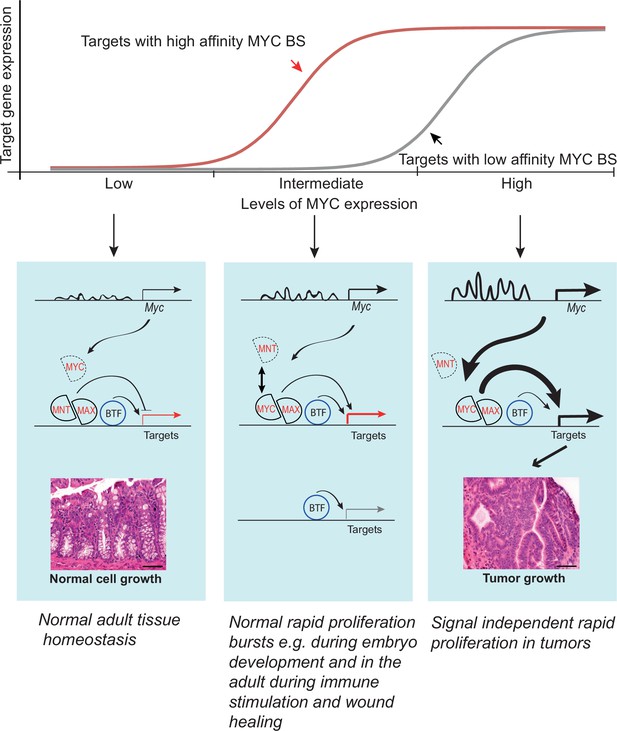
Model showing the activity of the Myc super-enhancer region during normal homeostasis (left) and cancer (right).
During normal tissue homeostasis (left), Myc enhancers are not strongly active, and MYC activity is relatively low. The MYC expression level is insufficient to recruit enough MAX proteins to MYC/MAX heterodimers to drive strong induction of the MYC target genes, which instead remain under the control of basal transcription factors (BTF). Under conditions of normal rapid proliferation as seen during embryonic development or during pathological insults in the adult, MYC is expressed at intermediate levels to elicit response from targets with high affinity binding sites (red). In cancer cells or cells grown in culture (right), upstream regulators such as Tcf7l2 and β-catenin activate the Myc super-enhancers, driving high levels of MYC expression. This leads to the formation of MYC/MAX heterodimers that strongly activate transcription of MYC target genes driving cancer cell growth. The high levels of MYC are also sufficient to induce target genes that harbor low affinity MYC binding sites (grey). The model is consistent with the model of Lorenzin et al (Lorenzin et al., 2016) who showed genes differ in their response to MYC levels due to differences in the MYC affinity of their promoters. Given that Myc super-enhancer region is tumor-specific, and induction of the MYC target genes are not required for normal homeostasis, it provides a promising target for antineoplastic therapies.
Additional files
-
Supplementary file 1
List of genes that show significant differential expression in the wild-type and the Myc△2–540/△2–540 colon (q-value <0.05).
- https://doi.org/10.7554/eLife.23382.018
-
Supplementary file 2
List of primers and guide RNA sequences used in this study.
- https://doi.org/10.7554/eLife.23382.019





