Negative regulation of urokinase receptor activity by a GPI-specific phospholipase C in breast cancer cells
Figures
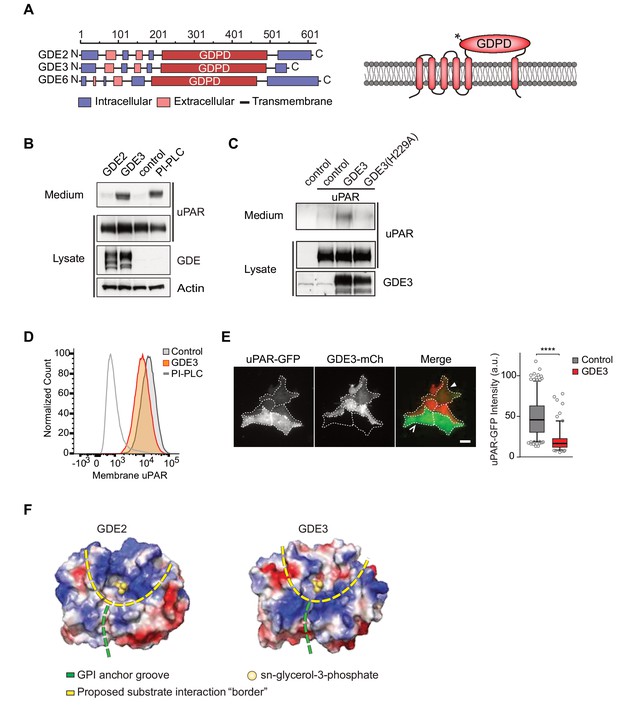
GDE3, but not GDE2, sheds uPAR from HEK293 cells.
(A) Domain structure of GDE2, GDE3 and GDE6 (left panel), and the transmembrane scheme of GDE3 (right panel). GDPD denotes catalytic glycerophosphodiesterase domain. Asterisk in GDPD domain depicts catalytic His residue in both GDE2 and GDE3. (B) Immunoblot analysis of uPAR release into the medium. HEK-uPAR cells transfected with empty vector (control), GDE2 or GDE3. PI-PLC served as positive control. (C) Mutant GDE3(H229A) fails to release uPAR. (D) Partial loss of uPAR from the plasma membrane by GDE3, as measured by flow cytometry. (E) TIRF microscopy reveals loss of uPAR from the basolateral plasma membrane. Box plot shows uPAR-GFP intensity at the ventral membrane (n = 3, mean ±SEM ****p<0001). (F) Homology modeling of the GDE2 and GDE3 catalytic domains showing surface charge distributions (blue, positive; red, negative; green line, putative GPI-binding groove; yellow line, proposed substrate-binding surface). The active site is indicated by glycerol-3-phosphate located at the template structure.
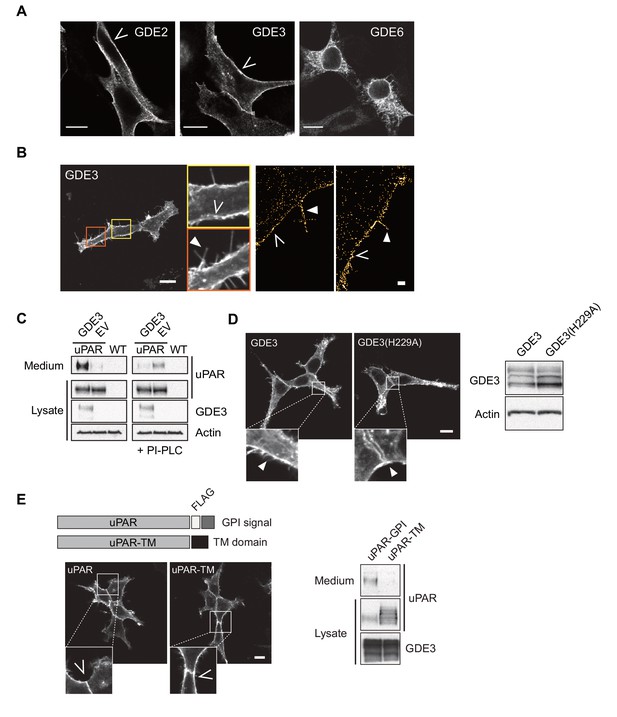
GDE subcellular localization and induction of uPAR release from HEK293-uPAR cells.
(A) Confocal images of HEK293 cells expressing human GDE2-HA, GDE3-HA or GDE6-HA, as indicated; bar, 10 μm. (B) GDE3 localizes to distinct microdomains (yellow square, open arrows) and filopodia-like extensions (orange square, solid arrow), as visualized by confocal and super-resolution microscopy. Bars, 10 μm and 1 μm respectively. (C) Immunoblot analysis showing that GDE3 reduces the membrane-anchored uPAR pool and competes with PI-PLC. The medium and lysates of HEK-uPAR cells expressing GDE3 or empty vector (EV) were analyzed without or with PI-PLC treatment (45 min.) of the cells, as indicated (left and right panels, respectively). Right lanes refer to wild-type (WT) HEK293 cells. (D) Expression and localization of GDE3 and mutant GDE3(H229A), as measured by immunoblotting and confocal microscopy, respectively, at 24 hr after transfection. Actin was used as a loading control. (E) uPAR-TM containing the transmembrane domain of EGFR is not released by GDE3, as shown by immunoblotting using anti-uPAR antibody, while uPAR-TM is properly expressed at the plasma membrane (bar, 10 μm).
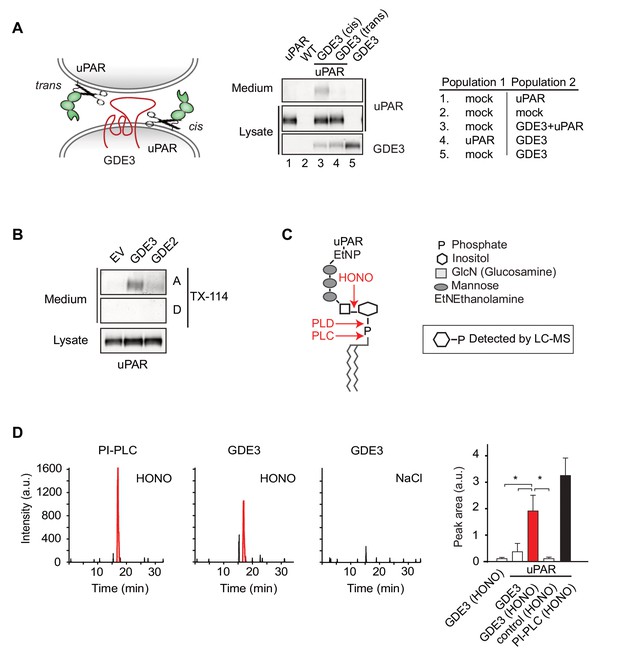
GDE3 is a GPI-specific PLC that cleaves uPAR in cis.
(A) (Left) Scheme showing uPAR cleavage in cis or trans. (Right) GDE3-expressing HEK-uPAR cells were mixed with a GDE3-deficient cell population, as indicated. Immunoblot analysis of uPAR in medium and cell lysates indicates that GDE3 acts in cis; mock refers to empty vector-transfected cells. (B) GDE3 expression leads to increased GPI-free suPAR. Conditioned medium from HEK-uPAR cells expressing GDE2 or GDE3 was subjected to Triton X-114 phase separation. suPAR in the aqueous (A) and detergent (D) fractions was analyzed by immunoblotting. (C) GPI-anchor with phospholipase cleavage sites indicated; HONO, nitrous acid. (D) Representative LC-MS ion chromatograms (m/z 259.02–259.03); inositol 1-phosphate peaks in red. HONO-treated suPAR contains inositol 1-phosphate (n = 3, mean ±SEM; *p<0.05).
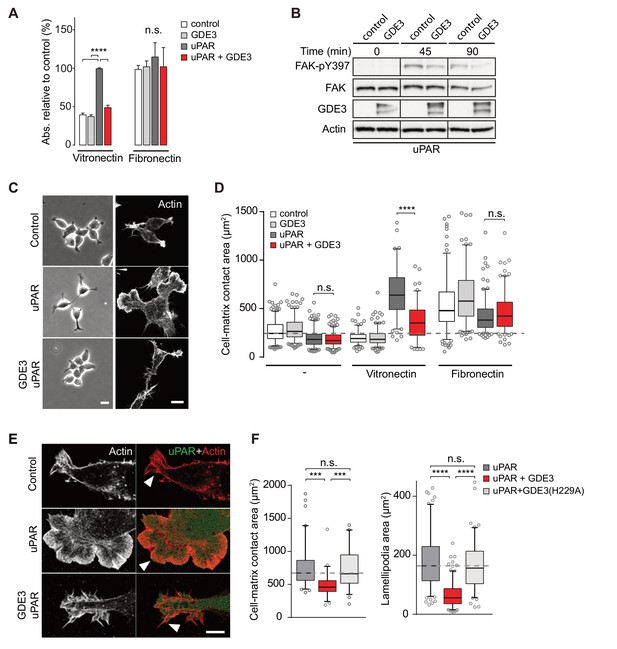
GDE3 suppresses uPAR activity in HEK-uPAR cells.
(A) uPAR confers increased adhesion to vitronectin, but not fibronectin, which is prevented by GDE3 expression (n = 3, mean ±SEM), ****p<0.0001; n.s., not significant. (B) GDE3 inhibits FAK activation during cell adhesion. FAK activity was assayed at the indicated times after plating. (C) GDE3 inhibits uPAR-induced cell spreading, scattering and lamellipodia formation on vitronectin (bar, 10 μm). (D) Cell-matrix contact area of cells expressing the indicated constructs plated on either fibronectin or vitronectin; (-) denotes cells on uncoated cover slips. Box plots show the mean of three independent experiments. Dotted line represents the mean of control cells (n = 3, mean ±SEM, ****p<0.0001; n.s., not significant). (E) uPAR-induced lamellipodia on vitronectin disappear upon GDE3 expression, as shown by super-resolution microscopy (bar, 5 μm). (F) Quantification of cell-matrix contact area in GDE3- and GDE3(H229A)-expressing cells. GDE3(H229A) fails to affect lamellipodia formation (n = 3, mean ±SEM ***p<0.001; ****p<0.0001).
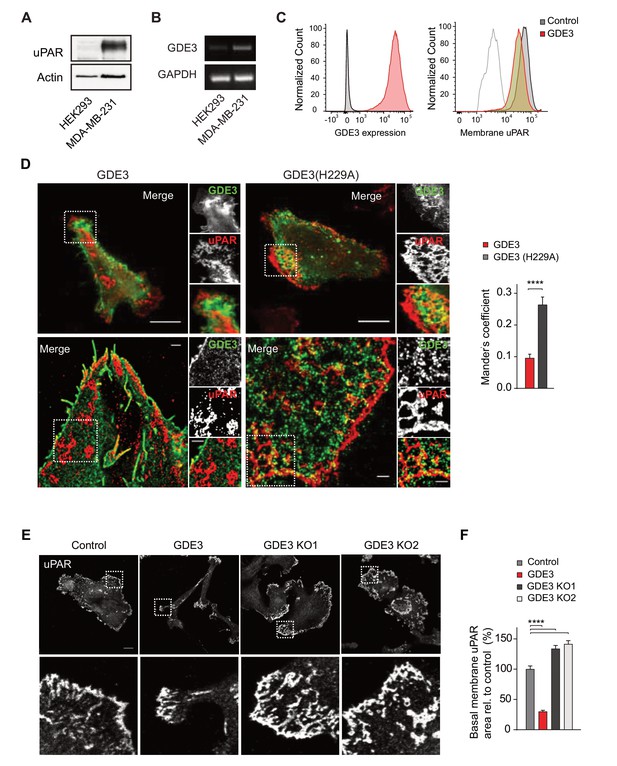
GDE3 depletes uPAR from distinct basolateral membrane domains in MDA-MB-231 breast cancer cells.
(A) Endogenous uPAR expression in MDA-MB-231 versus HEK293 cells, as determined by immunoblot. (B) Endogenous GDPD2 expression, as determined by qPCR analysis. (C) (left) Cell-surface expression of GDE3-mCherry of MDA-MB-231 cells expressing GDE3, as detected by flow cytometry. (Right) Cell-surface expression of uPAR in control (grey) and GDE3-expressing MDA-MB-231 cells (red), as detected by flow cytometry. (D) Confocal (top) and dual-color super-resolution microscopy images (bottom) of MDA-MB-231 cells expressing GDE3-GFP or catalytically dead GDE3(H229A)-GFP. Endogenous uPAR was immunostained in red. Merged images show colocalization of uPAR with GDE3(H229A) but not with wild-type GDE3 and uPAR. Scale bars, 10 μm (confocal) and 1 μm (super-resolution). Co-localization analysis (Mander's coefficient) on peripheral uPAR patches in confocal images was done using ImageJ software (n = 30 cells, three independent experiments). (E) Endogenous uPAR staining in control, GDE3-overexpressing and GDE3 knockout MDA-MB-231 cells plated on vitronectin. Two distinct GDE3 knockout clones (KO1 and KO2) were examined, as indicated. Scale bar,10 μm. (F) Quantification of basolateral uPAR-containing membrane domains referring to the cells in panel (E) (n = 3, mean ±SEM, ****p<0.0001). GDE3 suppresses the vitronectin- and uPAR-dependent phenotype of MDA-MB-231 breast cancer cells.
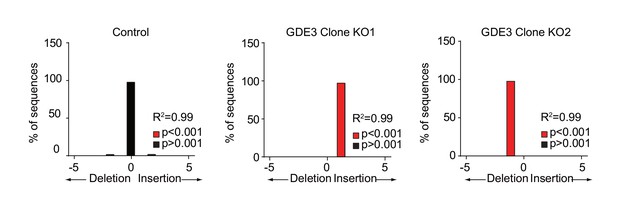
GDE3 knockout validation.
GDE3 knockout in MDA-MB231 cells was achieved using CRISPR/Cas9 genome editing. Surviving colonies were screened for cassette integration and indels into the query gene by PCR. Two genetically individual clones (KO1 and KO2) were selected, and knockout was confirmed using Sanger sequencing followed by TIDE deconvolution (Brinkman et al., 2014).
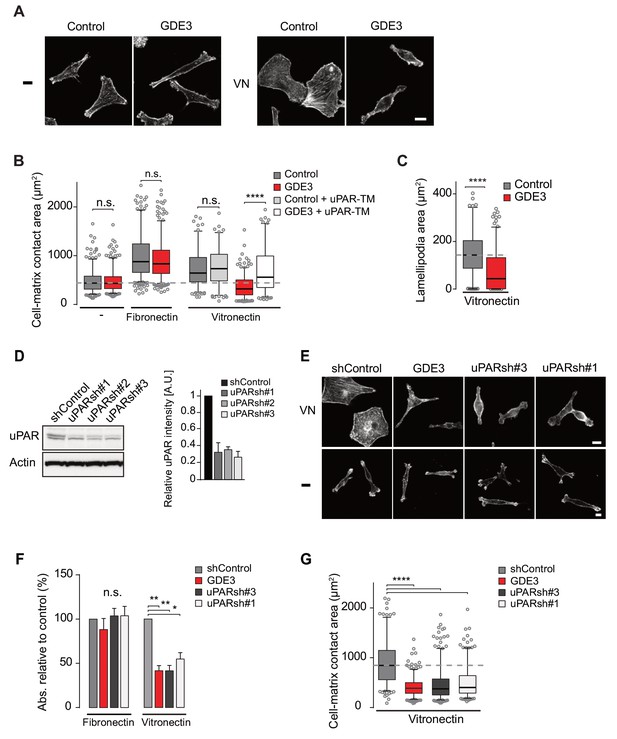
GDE3 suppressess the vitronectin- and uPAR-dependent transformed phenotype of MDA-MB-231 breast cancer cells.
A) Confocal images showing that GDE3 prevents cell spreading and lamellipodia formation on vitronectin (VN) but not on uncoated cover slips (-). Bar, 10 μm. (B) Quantification of reduced cell spreading on vitronectin by GDE3. Non-cleavable uPAR-TM prevents GDE3 attack. ****p<0.0001; n.s., not significant. (C) Quantification of lamellipodia formation on vitronectin. ****p<0.0001. (D) Immunoblot analysis of shRNA-mediated uPAR knockdown; maximum knockdown was achieved by small hairpins #1 and #3. The upper protein band represents full-length uPAR, the lower band proteolytically cleaved uPAR(D2 +D3) (Høyer-Hansen et al., 1992). (E) GDE3 overexpression mimics the uPAR knockdown phenotype in cells plated on vitronectin (VN); bar, 10 μm. (-) denotes cells on non-coated cover slips. (F,G) Quantification of cell adhesion (F) n = 3; mean ±SEM) and cell spreading (G) induced by GDE3 and uPAR knockdown on the indicated substrates. *p<0.05 **p<0.01; ****p<0.0001. GDE3 overexpression attenuates the uPAR-dependent transformed phenotype of breast cancer cells.
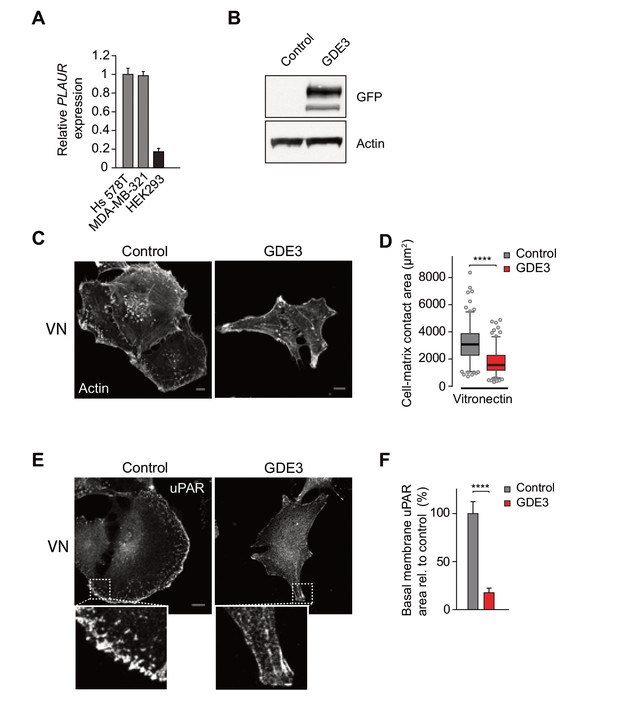
GDE3 overexpression suppresses the uPAR-vitronectin-dependent phenotype in Hs578T breast cancer cells.
(A) Relative expression of uPAR (encoded by PLAUR)) in the indicated cell lines, as determined by qPCR. (B) GDE3-GFP expression in Hs578T cells, using anti-GFP antibody. (C) Confocal images showing cell morphology (F-actin staining) of GDE3-expressing versus control cells plated on vitronectin (Bar, 10 μm). (D) Quantification of cell area of GDE3-expressing versus control Hs578Tcells, plated on vitronectin (n = 3, mean ±SEM, ****p<0.0001). (E) Hs578T cells with or without stable GDE3-GFP expression were plated on vitronectin and stained for endogenous uPAR. Representative images of the basolateral membrane are depicted. Bar, 10 μm. (F) uPAR in the basolateral membrane was quantified in control and GDE3-expressing Hs578T cells plated on vitronectin (n = 3, mean ±SEM, ****p<0.0001).
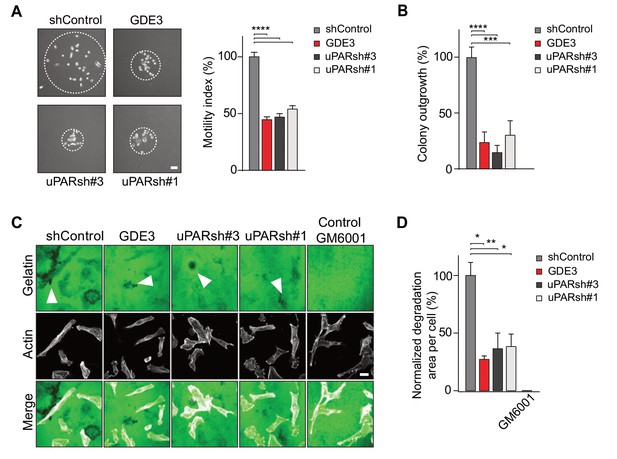
GDE3 attenuates the uPAR-dependent transformed phenotype of breast cancer cells.
(A) GDE3 overexpression in MDA-MB-231 cells mimics uPAR knockdown in inhibiting cell scattering. MDA-MB-231 cells grow out as scattered colonies; bar, 100 μm (n = 3, mean ±SEM; ***p<0.001; ****p<0.0001). (B) GDE3 overexpression mimics uPAR knockdown in suppressing colony formation. colonies were counted after 14 days (n = 3, mean ±SEM; ***p<0.001; ****p<0.0001. (C) Degradation of a gelatin matrix mixed with vitronectin by MDA-MB-231 cells in the presence of serum (bar, 10 μm). Arrows point to black spots where gelatin was degraded. Metalloprotease inhibitor GM6001 was used as a control. (D) Quantification of matrix degradation at 20 hr after plating (mean ±SEM; *p<0.05; **p<0.01).
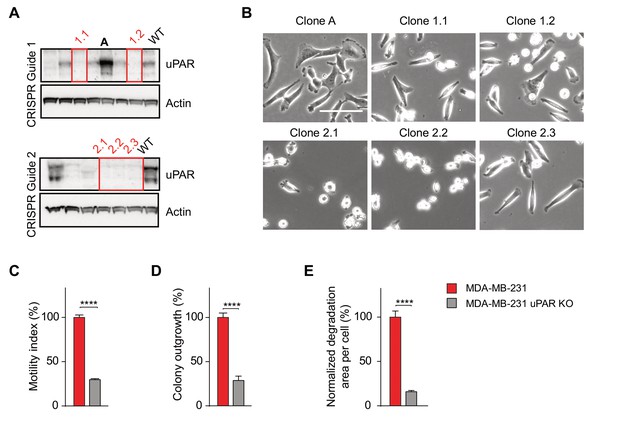
uPAR knockout in MDA-MB-231 cells phenocopies GDE3 overexpression.
(A) uPAR knockout in MDA-MB-231 cells was achieved using CRISPR/Cas9 genome editing. uPAR knockout was assayed by immunoblot analysis using anti-uPAR antibody. Individual clones were picked and plated on vitronectin. (B) uPAR-deficient MDA-MB-231 cells show GDE3 overexpression phenotype. uPAR KO cells show decreased cell spreading and lamellipodia formation on vitronectin. scattering (bar, 100 μm). (C) uPAR-deficient MDA-MB-231 cells show reduced cell motility (n = 3, mean ±SEM; ****p<0.0001). (D) MDA-MB-231 wild-type or uPAR knockout colonies were counted after 14 days. ****p<0.001. (E) Gelatin matrix degradation by wild-type and uPAR-deficient MDA-MB-231. Degradation was quantified at 20 hr after plating (mean ±SEM; ****p<0.0001).
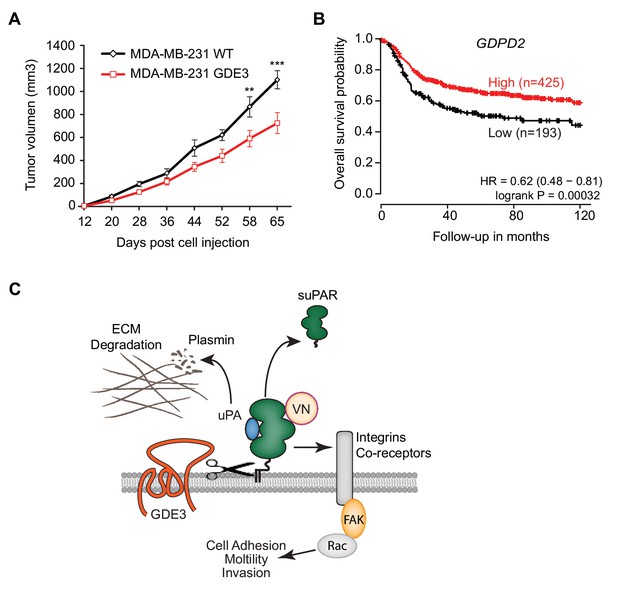
GDE3 overexpression in breast cancer cells slows tumor growth in mice, correlating with higher survival probability in patients.
(A) Female nude mice (n = 16) were injected with either wild-type or GDE3-overexpressing MDA-MB-231 cells into the mammary fat pad, as described under Materials and methods. Tumor volume was measured every three days after injection for 9 weeks until the tumor had grown to the appropriated size (data represent the mean ±SEM; **p<0.01; ***p<0.001, t-test corrected for multiple comparison). (B) High GDPD2 expression significantly correlates with higher survival rate in triple-negative (basal-like) subtype breast cancer patients (N = 618). HR, hazard ratio with 95% confidence interval indicated. Analysis based on microarray data (www.kmplot.com). (C) Schematic model. GDE3 functions as a GPI-PLC that cleaves and sheds uPAR, leading to loss of uPAR function. VN, vitronectin, FAK, focal adhesion kinase, suPAR, soluble uPAR.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.23649.014





