Poly(A) tail length regulates PABPC1 expression to tune translation in the heart
Figures
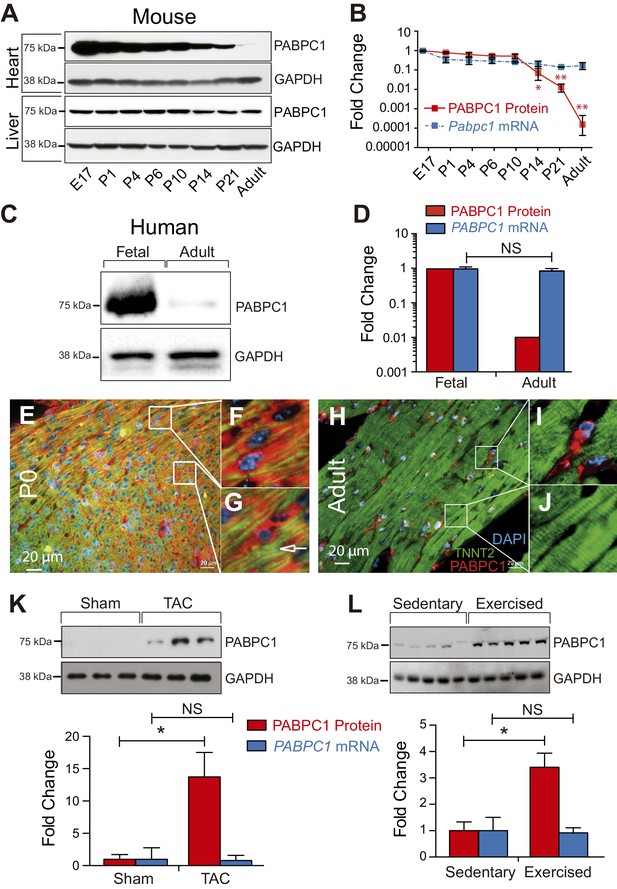
PABPC1 is dynamically regulated during cardiac development and hypertrophy.
(A–D) Relative quantification of PABPC1 protein (immunoblots) and mRNA (qPCR) levels normalized to GAPDH during mouse heart and liver development (A, B) and in human fetal and adult hearts (C, D). (E–J) Immunofluorescent images of mouse postnatal day 0 (P0) and 8-week-old adult hearts stained for PABPC1 (red), cardiac troponinT (green), and DAPI (blue). Insets G and J show cardiomyocytes, while F and I show non-cardiomyocytes. Immunoblots and quantification of PABPC1 protein and mRNA from wild-type mouse hearts 8 weeks after (K) thoracic aortic constriction (TAC) or (L) 10-week exercise training. Data are mean ± s.d (n = 3); *p<0.05, unpaired two-tailed t-test. NS, not significant.
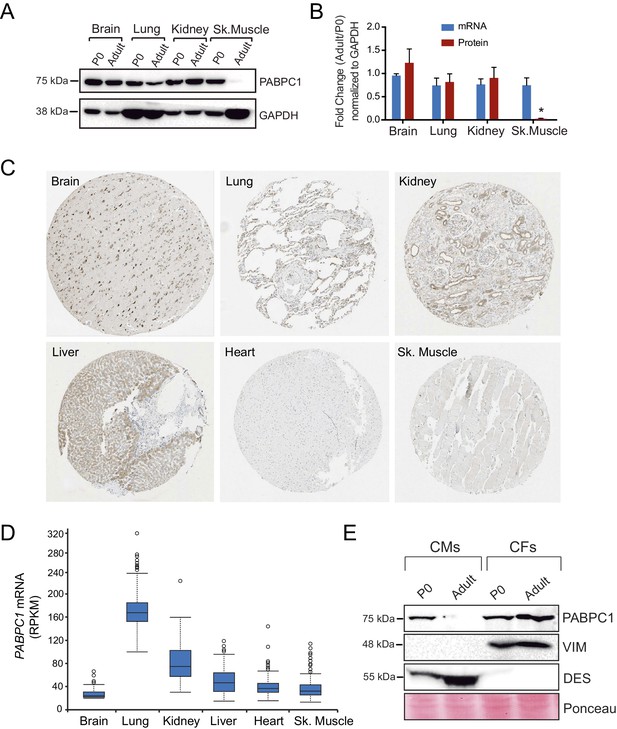
Post-transcriptional silencing of PABPC1 is muscle-specific.
(A) Immunoblots for PABPC1 from postnatal day 0 (P0) and 8-week-old adult mice from the indicated tissue with GAPDH as a loading control. (B) Quantification of PABPC1 protein and mRNA expression (qPCR). Data are mean ± s.d (n = 3); *p<0.005 unpaired two-tailed t-test. (C) PABPC1 immuno-histochemistry images based on anti-PABPC1 antibody (Abcam ab21060) from the Human Protein Atlas Database (www.proteinatlas.org) showing that PABPC1 is abundantly expressed in other adult tissues but not heart and skeletal muscle. (D) RPKM values from the Human Protein Atlas Database showing relative Pabpc1 mRNA levels in various human tissues. (E) Western blot of purified P0 and adult cardiomyocytes (CMs) and cardiac fibroblasts (CFs). Blotting for Desmin (DES, CM marker) and Vimentin (VIM, CF marker) show clean separation of cell types.
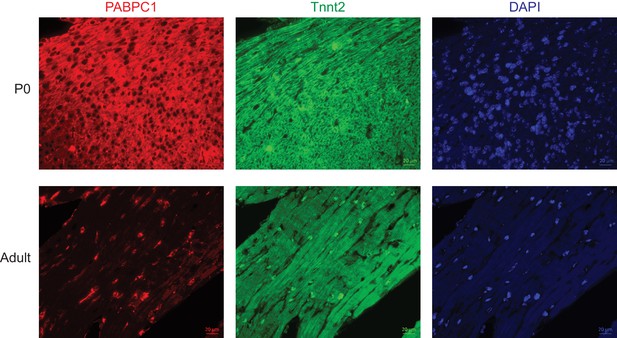
Post-transcriptional silencing of PABPC1 is muscle-specific.
Single-channel immunofluorescent images of mouse postnatal day 0 (P0) and 8-week-old adult hearts stained for PABPC1 (red), cardiac troponinT (green), and DAPI (blue).
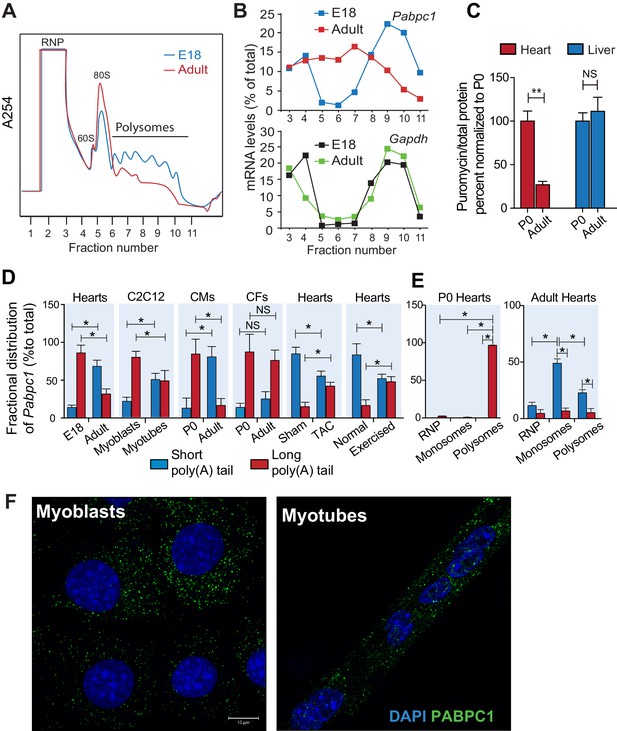
Poly(A) tail length determines cell-type and developmental stage-specific translation of PABPC1.
(A). Polysome profile of embryonic day 18 (E18) and adult mouse hearts. (B) Percentage of Pabpc1 and Gapdh mRNAs measured by qPCR in each fraction collected from the polysome profiling. (C) Neonatal and 8-week-old adult wild-type mice were pulsed with puromycin through an intraperitoneal injection. Forty-five minutes following injection, heart and liver tissues were harvested for immunoblotting with anti-puromycin antibody. De novo protein synthesis was quantified as the ratio of puromycin labeled peptides to total protein. (D) Fractional distribution of Pabpc1 mRNAs with short and long poly(A) tails in whole heart, C2C12 cells, cardiomyocytes (CMs), cardiac fibroblasts (CFs), whole heart after TAC surgery, and whole heart after exercise (measured by qPCR following poly(A) tail fractionation). (E) Poly(A) tail length status of Pabpc1 mRNA within P0 and adult heart RNP, monosome, and polysome fractions from sucrose gradients. (F) Pabpc1 single-molecule RNA-FISH in C2C12 myoblasts and myotubes. Data are mean ± s.d (n = 3); *p<0.05, **p<0.005 unpaired two-tailed t-test; NS, not significant.
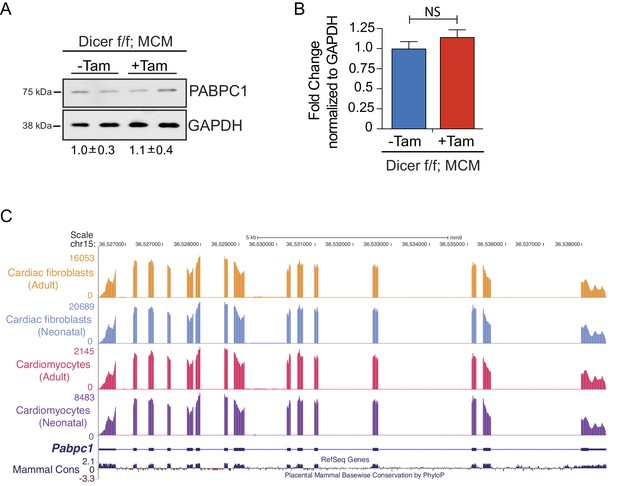
Regulation of PABPC1 expression in adult heart is independent of miRNAs or alternative splicing.
Targeted deletion of Dicer in adult cardiomyocytes was obtained by treating 8-week-old Dicer f/f; MCM mice with tamoxifen (20 mg/Kg/day) for 5 consecutive days21. Forty-eight hours after the last injection, heart tissues were harvested. (A) Relative PABPC1 protein (immunoblots) or (B) mRNA (qPCR) levels were measured in the indicated samples. Fold change in PABPC1 protein was determined by relative quantification of band intensities, normalized to GAPDH (shown below the blots). Data are mean ± s.d (n = 3); unpaired two-tailed t-test. NS, not significant. (C) UCSC genome browser tracks of Pabpc1 mRNA in cardiomyocytes and fibroblasts from neonatal and adult mice16.
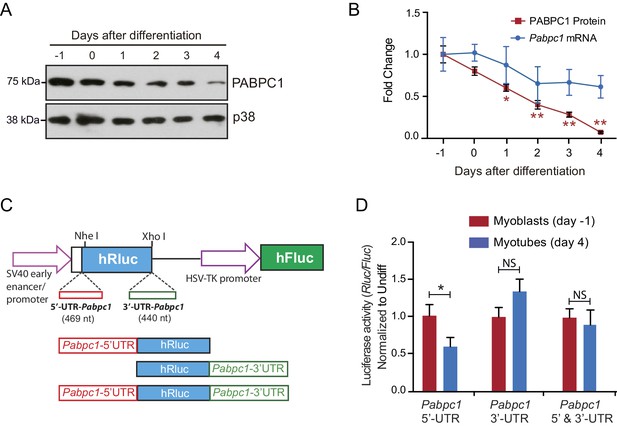
Limited influence for 5’ and 3’ untranslated regions (UTRs) of Pabpc1 on luciferase protein translation during C2C12 differentiation.
(A) Representative immunoblot demonstrating a steady decrease in PABPC1 protein levels during C2C12 differentiation; p38 was used as a loading control. (B) Quantification of PABPC1 protein (immunoblots) and mRNA (qPCR) levels relative to p38 during C2C12 differentiation. (C) Schematic of the reporters designed to assess the effect of Pabpc1 5’ and 3’ UTRs on luciferase protein expression. (D) Relative luciferase activity derived from C2C12 cells transfected with different reporter constructs shows that only Pabpc1 5’ UTR has a moderate effect (twofold) on the reporter activity in differentiated myotubes, whereas the 3’ UTR and the combination of UTRs have no effects. Data are mean ± s.d (n = 3); *p<0.05, unpaired two-tailed t-test. NS, not significant.
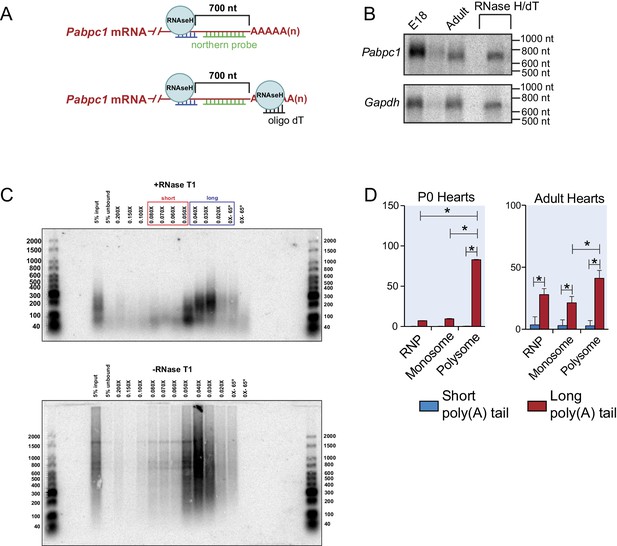
Experimental design of northern blot and RNA isolation based on poly(A) tail length through gradient purification.
(A) Oligo design for the Pabpc1 northern assay. Oligos were designed for the Pabpc1 mRNA to be targeted in an RNAseH digestion leaving 700nt downstream of the cleavage site and the native poly(A) tail. (B) Northern blot using a radiolabeled probe against Pabpc1 from E18 and 8-week-old adult mouse hearts following RNAseH cleavage. In the final lane, oligo dT was also added to the RNAseH reaction so that the Pabpc1 transcript was cleaved at both ends, leaving just the 700nt region. A similar RNAseH cleavage assay was used for GAPDH as a control. (C) Adult mouse heart total RNAs with and without RNAse T1 treatment were mixed with biotinylated oligo(dT) and bound to the streptavidin-conjugated beads. Different salt concentrations were used to elute mRNAs. The differing poly(A) tail lengths of eluted RNAs were checked by northern blot analysis using an oligo-dT40 probe. This served as optimizations for further purifications of short and long poly(A) tail RNAs that were subjected to qPCR analysis using gene-specific primers. (D) Poly(A) tail status of P0 and Adult Gapdh in polysome gradient fractions as a control experiment. Gapdh is enriched in the long-tailed and polysome fractions as expected.
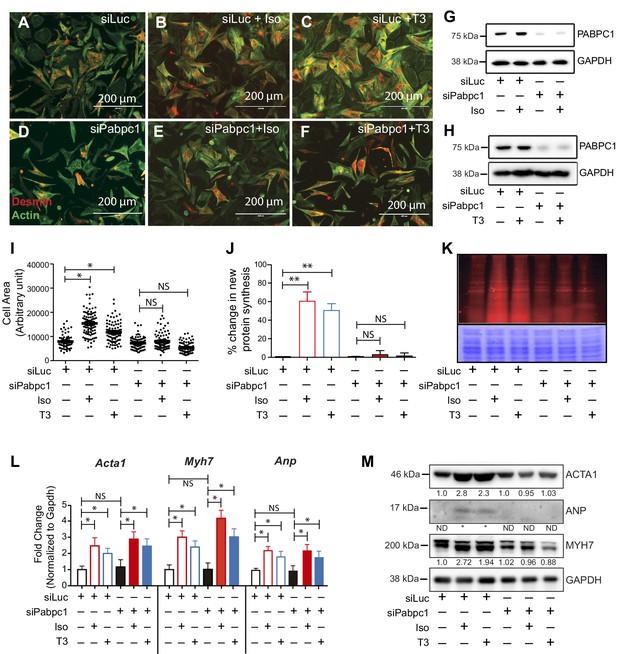
Knockdown of PABPC1 in neonatal mouse cardiomyocytes prevents stimulus-induced hypertrophy and protein synthesis.
(A–F) Primary cardiomyocytes isolated from newborn mice were transfected with siRNA against control Luciferase or Pabpc1. Twelve hours following transfection, cells were treated with isoproterenol (Iso) or triiodothyronine (T3) for 36 hr to induce hypertrophy. Cells were stained for Desmin and Actin by immunofluorescence to verify cardiomyocyte identity and measure cell area. (G, H) Immunoblots demonstrating efficient PABPC1 knockdown 48 hr after siRNA treatments with either Iso or T3. (I) Quantification of cell area 36 hr post Iso or T3 treatments. (J, K) Measurement of new protein synthesis using Click-iT homopropargylglycine assay after 2 hr of Iso or T3 treatments. (L) Quantification of mRNA (qPCR) from neonatal cardiomyocytes for each condition shows significant upregulation of mRNA for Acta1, Myh7, and Anp in response to Iso or T3 treatments. (M) Representative immunoblot showing that protein levels of ACTA1, MYH7, and ANP are increased after Iso or T3 treatments in the control Luciferase knockdown but synthesis is prevented when Pabpc1 is knocked down. Data are mean ± s.d (n = 3); *p<0.05, **p<0.01, one-way analysis of variance (ANOVA) plus Dunnett’s post-hoc test. NS, not significant.
-
Figure 3—source data 1
Source data for cell area of cultured neonatal cardiomyocytes treated with siRNA and either Iso or T3.
- https://doi.org/10.7554/eLife.24139.011
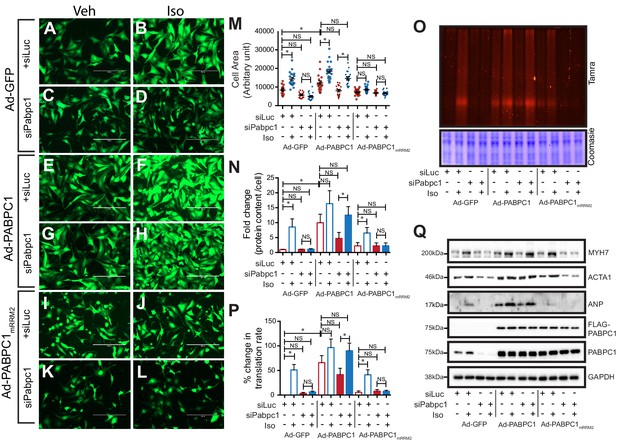
PABPC1–eIF4G1 interactions control stimulus-induced new protein synthesis and hypertrophy.
(A–L) Representative images of neonatal cardiomyocytes infected with adenovirus expressing GFP, wildtype PABPC1, or a PABPC1 RRM2 mutant (that does not interact with eIF4G1), transfected with siRNA against endogenous Pabpc1 or Luciferase, and treated with isoproterenol (Iso) or vehicle (Veh). Quantification of (M) cell areas, (N) total protein content, (O, P) rate of new protein synthesis measured by Click-iT homopropargylglycine fluorescence assay after respective treatments. (Q) Representative immunoblots of hypertrophy markers. Data are mean ± s.d (n = 3); *p<0.05, one-way analysis of variance (ANOVA) plus Dunnett’s post-hoc test. NS, not significant.
-
Figure 4—source data 1
Source data for cell area of cultured neonatal cardiomyocytes after treatment with siRNA, adenovirus, and Iso.
- https://doi.org/10.7554/eLife.24139.013
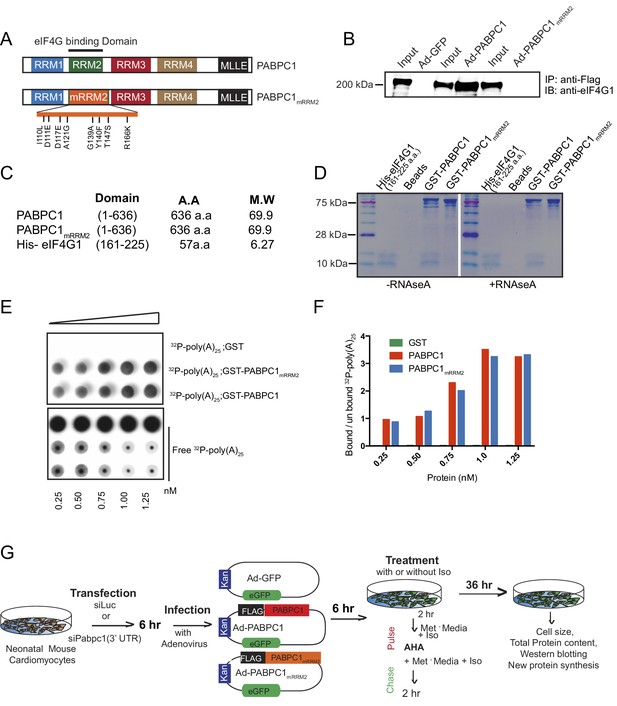
PABPC1mRRM2 can bind to poly(A) RNA but does not interact with eIF4G1.
(A) Schematic of the PABPC1 protein with key domains labeled. Mutations of the RRM2 domain (PABPC1mRRM2) with eight amino acid replacements to interrupt PABPC1-eIF4G binding were made using the available structural information5. (B) C2C12 myoblasts were infected with FLAG-PABPC1, FLAG-PABPC1mRRM2, and control GFP adenoviruses followed immunoprecipitation by anti-FLAG antibody and blotting for endogenous eIF4G1. FLAG-PABPC1 construct efficiently immunoprecipitated eIF4G1, whereas the negative control FLAG-GFP and the PABPC1mRRM2 did not. (C) Constructs used for bacterial expression and purification for mouse PABPC1, PABPC1mRRM2, and a PABP-interacting fragment of mouse eIF4G15. (D) Equimolar amounts of purified His-eIF4GI 61-225 and GST-PABPC1 or GST-PABPC1mRRM2 were mixed and incubated with or without RNase A. Protein complexes were isolated using Glutathione-Magnetic beads, separated on 10% SDS-PAGE gel and visualized by Coomassie staining. GST-PABPC1 showed binding to His-eIF4GI 61-225 whereas GST-PABPC1mRRM2 or empty beads did not. (E) A gradient of concentrations of GST, GST-PABPC1mRRM2, and GST-PABPC1 along with 32P labeled poly(A)25 oligo were used to capture Protein-RNA probe complexes and un-bound RNA probes respectively in a filter binding assay. Following incubation, radioactive signal was measured to quantify how much 32P-poly(A)25 remained bound to the protein and how much washed through to the nucleic acid membrane. GST did not bind any 32P-poly(A)25. Both wild-type and mutant PABPC1mRRM2 were capable of binding the 32P-poly(A)25. (F) Quantification of the Geiger counts on the protein membrane. (G) Schematic depicting the experimental set up of PABPC1 rescue experiments in neonatal cardiomyocytes.
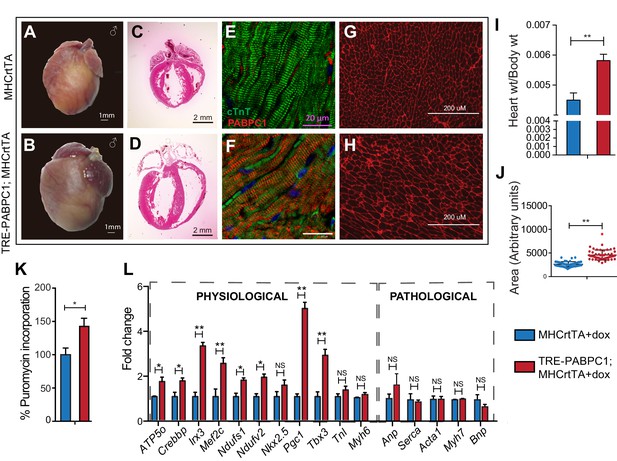
Forced expression of PABPC1 in adult cardiomyocytes induces physiologic hypertrophy.
(A–H) Representative whole heart, H&E, immunofluorescent, and WGA-stained sections of 2-week doxycycline (Dox)-induced MHCrtTA transgenic controls and TRE-PABPC1; MHCrtTA bitransgenic mice. (I) Heart-to-body weight ratios (n = 6). (J) Cell area quantified from WGA-stained sections (n = 3). (K) Global rate of translation based on puromycin incorporation in hearts of injected mice (n = 6). (L) Relative mRNA levels of indicated physiological and pathological hypertrophy markers normalized to GAPDH (qPCR, n = 9). Data are mean ± s.d; *p<0.05 unpaired two-tailed t-test. NS, not significant.
-
Figure 5—source data 1
Source data for cardiomyocyte areas performed on WGA stained heart tissue sections of 2-week doxycycline-induced MHCrtTA transgenic controls and TRE-PABPC1; MHCrtTA bitransgenic mice.
- https://doi.org/10.7554/eLife.24139.016
-
Figure 5—source data 2
Source data for Figure 5—figure supplement 1.
Sheet 1: Source data for exercise performance between 6-month doxycycline induced MHCrtTA transgenic controls and TRE-PABPC1; MHCrtTA bitransgenic mice. Sheet 2: Source data for cardiac function tests between 6-month doxycycline-induced MHCrtTA transgenic controls and TRE-PABPC1; MHCrtTA bitransgenic mice.
- https://doi.org/10.7554/eLife.24139.017
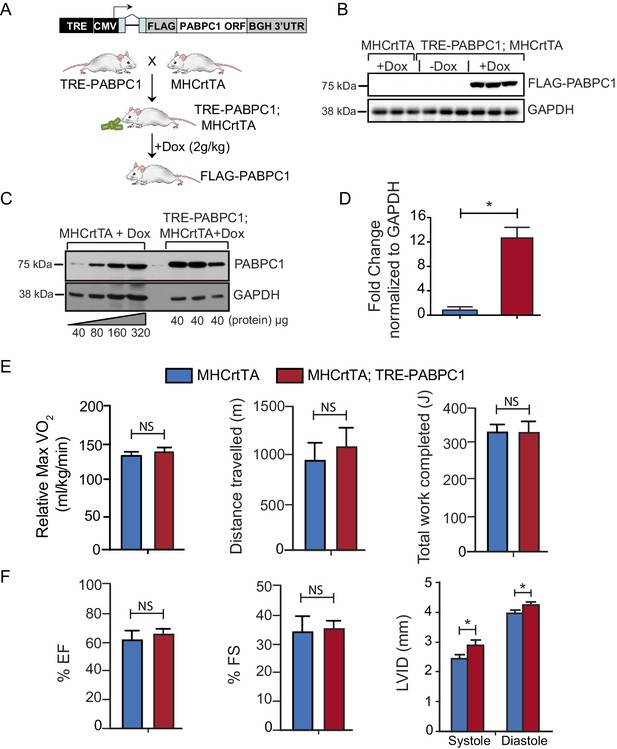
Generation of tetracycline-inducible, heart-specific PABPC1 transgenic mouse model.
(A) The TRE-PABPC1 construct expresses mouse PABPC1 containing an N-terminal Flag tag driven by a TRE and a CMV minimal promoter. TRE-PABPC1 mice were mated with Myh6-rtTA (MHCrtTA) mice to generate TRE-PABPC1; MHCrtTA bitransgenic mice as shown. Eight-week-old adult bitransgenic and MHCrtTA control animals were fed 2 g/kg doxycycline (Dox) containing diet for 2 weeks to induce FLAG-PABPC1 expression specifically in cardiomyocytes. (B) Immunoblot against FLAG shows that exogenous PABPC1 in the heart is only expressed when Dox is present in the diet. (C) Immunoblot against PABPC1 shows the relative amount of induction relative to the wild-type mice. (D) Quantification of the blot in C Data are mean ± s.d (n = 6); *p<0.05, unpaired two-tailed t-test. (E) MHCrtTA control and bitransgenic mice were induced with 2 g/kg Dox diet for 6 months and then subjected to acute exercise performance test to measure effects of PABPC1 expression on relative Max VO2 consumption, distance traveled, or work completed. (F) Echocardiograms were performed to calculate ejection fraction (EJ), fractional shortening (FS), and left ventricular internal dimension at systole and diastole (LVID). Data are mean ± s.d (n = 6–12); *p<0.05, unpaired two-tailed t-test. NS, not significant.
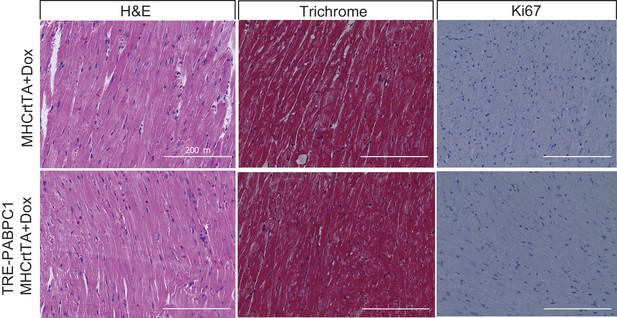
Induced expression of cardiomyocyte specific PABPC1 does not lead to damage.
Histological sections of 6-month 2 g/kg doxycycline (Dox)-induced MHCrtTA transgenic and TRE-PABPC1; MHCrtTA bitransgenic mice. Hematoxylin and eosin or trichrome staining shows normal cytoarchitecture of the heart, no myofiber disarray or presence of inflammatory cells, and no signs of damage or interstitial fibrosis in either genotype. Immunohistochemistry with Ki67 antibody shows absence of any proliferating cells with PABPC1 overexpression.
Additional files
-
Supplementary file 1
This document contains detailed information on tools used in the study. Sheet 1 contains the forward and reverse sequences of all primers organized by RT-qPCR, genotyping of mouse lines, cloning for plasmid constructs, RNAse H cleavage, and generation of northern blot probes. Sheet 2 contains antigen, manufacturer, product number, and RRID information for all antibodies used. Sheet 3 contains the probe sequences for Pabpc1 RNA-FISH.
- https://doi.org/10.7554/eLife.24139.020





