The murine catecholamine methyltransferase mTOMT is essential for mechanotransduction by cochlear hair cells
Figures
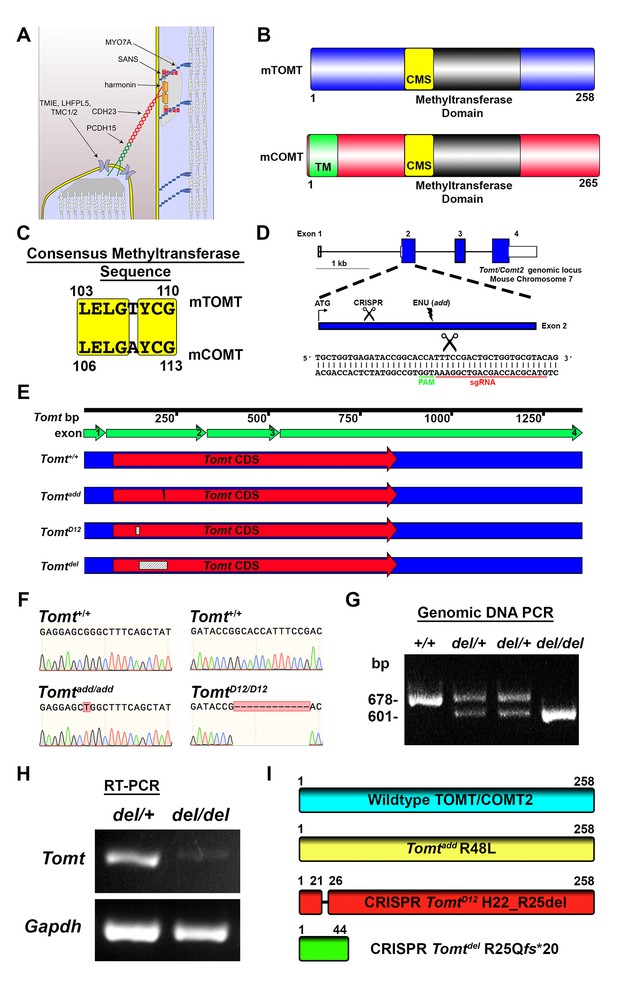
Generation of Tomt mutant mice.
(A) Schematic of the tip-link complex in hair cell stereocilia. (B) Schematic comparison of mouse TOMT (mTOMT, Uniprot AIY9I9) and mouse COMT (mCOMT, Uniprot O88587). Putative methyltransferase domain is indicated in black. Amino acid numbers are below. Consensus methyltransferase sequence (CMS, [Vidgren et al., 1994]) is depicted in yellow. Transmembrane domain (TM) from membrane-bound isoform of COMT is in green. (C) Conserved amino acid residues in consensus methyltransferase sequence of mTOMT and mCOMT. (D) Genomic locus of mouse Tomt (NCBI gene 791260), located on chromosome 7. Four Tomt exons are depicted as open boxes, with coding exons in blue. Introns are shown as solid black line. Exon 2, the first coding exon, is enlarged below, with relative locations indicated for ATG, clustered regularly interspaced short palindromic repeats (CRISPR) sgRNA recognition site, and add N-ethyl-N-nitrosourea (ENU) mutation site (Du et al., 2008). At bottom is Tomt exon 2 sequence showing CRISPR sgRNA and protospacer adjacent motif (PAM), and site of Cas9 cleavage (scissors). (E) Schematic of Tomt mRNA (NCBI NM_001081679.1) and location of mutations. Tomt exons are indicated with green arrows. Consensus Tomt coding sequence (CDS, NCBI CCDS40044.1) is in red. Location of add mutation is indicated with lightning bolt. Two unique deletions were identified in founder mice after CRISPR/Cas9 pronuclear injection of Tomt-specific sgRNA and Cas9. Genetic mouse lines were generated containing either a 12 base-pair (bp) deletion (TomtD12) or 77 bp deletion (Tomtdel) in Exon 2 of Tomt. Deletions are indicated as dashed-line boxes. (F) Genomic DNA sequencing results from Tomt+/+, Tomtadd/add, and TomtD12/D12 mice demonstrating mutations. (G) PCR of Tomt genomic DNA from Tomt+/+, Tomtdel/+, and Tomtdel/del demonstrating 77 bp deletion. (H) RT-PCR results for Tomt and Gapdh from inner ear tissue from Tomtdel/+ (+/−) and Tomtdel/del (−/−). (I) Predicted protein structure of wild-type and mutant TOMT. From top: wild-type TOMT; add mutation (R48L); CRISPR 12 bp in-frame deletion (TomtD12, H22_R25 del) leading to loss of four animo acids; and CRISPR 77 bp deletion (Tomtdel, R25Qfs*20) leading to a frame-shifted amino acid sequence, premature stop codon and truncated protein.
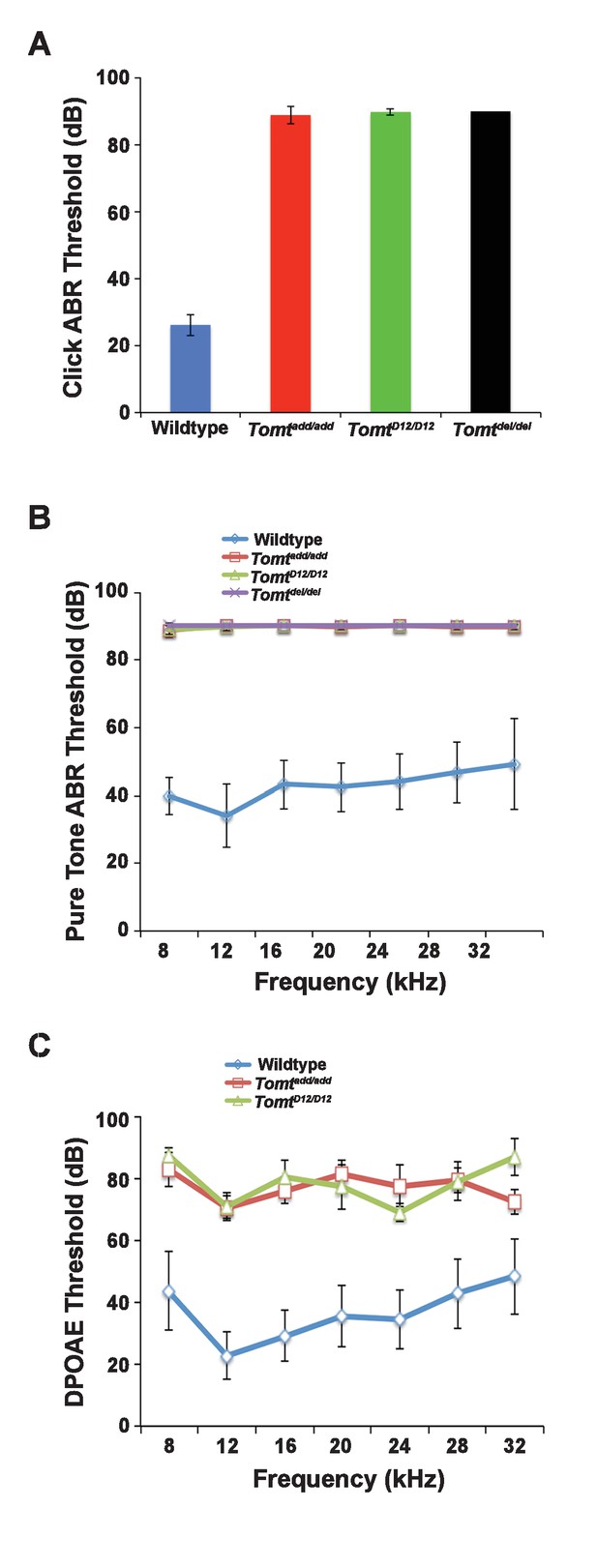
Hearing function.
(A) Auditory brainstem response (ABR) threshold values in response to Click stimuli from 3- to 5-week old wildtype (n = 13), add (Tomtadd/add, n = 10), H22_R25del (TomtD12/D12, n = 7), and R25Qfs (Tomtdel/del, n = 4). (B) ABR threshold values in response to pure tone stimuli of various frequencies for wildtype (n = 13), add (Tomtadd/add, n = 10), H22_R25del (TomtD12/D12, n = 7), and R25Qfs (Tomtdel/del, n = 4). (C) Threshold values for distortion product otoacoustic emission (DPOAE) stimuli presented at various frequencies for wildtype (n = 13), add (Tomtadd/add, n = 10) and H22_R25del (Tomt D12/D12, n = 7). All values are mean±SD.
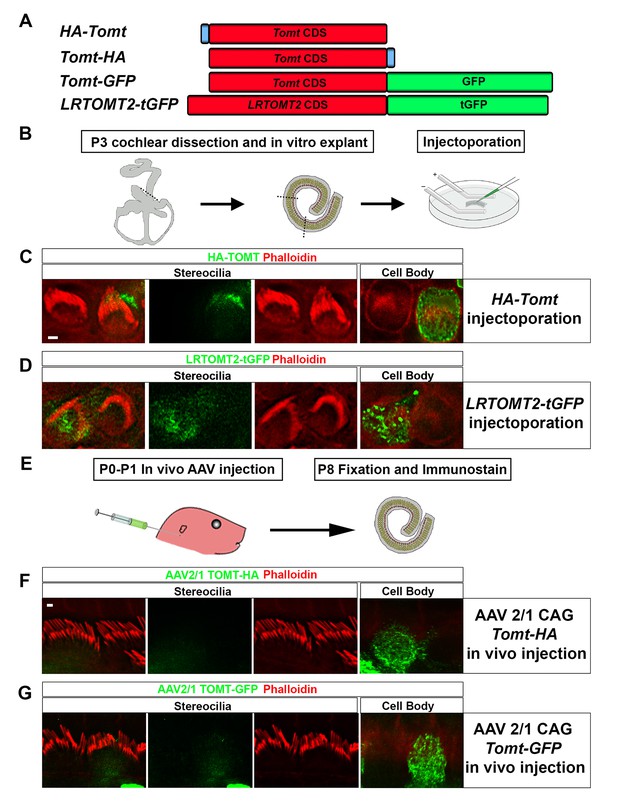
Localization of epitope-tagged TOMT in cochlear hair cells.
(A) Schematic of constructs used for localization studies. Constructs included Mouse TOMT coding sequence tagged at the N-terminus with HA (HA-Tomt), C-terminus with HA (Tomt-HA), and C-terminus with GFP (Tomt-GFP). LRTOMT2, the human homolog of mouse TOMT, was tagged on the C-terminus with tGFP (LRTOMT2-tGFP). (B) Depiction of experimental paradigm for injectoporation of epitope-tagged constructs (Xiong et al., 2014). The Organs of Corti of P3 mice were dissected, sectioned into three parts, and maintained as in vitro explants. Plasmid DNA was microinjected into the space surrounding the basal compartment of hair cells, followed by electroporation. Injectoporated explants were cultured for 24 hr, fixed, and immunostained. (C) OHCs injectoporated with HA-TOMT were stained with anti-HA antibody (green) and phalloidin (red) to label actin. Optical sections at the level of the stereocilia and cell body are shown. Note that HA-TOMT localizes to the cell body but not the stereocilia. (D) OHCs injectoporated with LRTOMT2-tGFP (green) were stained with phalloidin (red) to label actin. Optical sections at the level of the stereocilia and cell body are shown. Note that LRTOMT2-tGFP localizes to the cell body but not the stereocilia. (E) Schematic to describe in vivo inner ear injections of AAV serotype 2/1 viral vectors expressing TOMT-HA or TOMT-GFP from a CAG promoter. At P0-P1, AAV vectors were injected into the inner ear of pups using a Hamilton syringe with a beveled needle tip. One week later, inner ears were fixed and immunostained. (F) IHCs expressing AAV 2/1 CAG TOMT-HA, stained with anti-HA antibody (green) and phalloidin (red). Optical sections at the level of the stereocilia and cell body are shown. Note that TOMT-HA localizes to the cell body but not the stereocilia. (G) IHCs expressing AAV 2/1 CAG TOMT-GFP, stained with anti-GFP antibody (green) and phalloidin (red). Optical sections at the level of the stereocilia and cell body are shown. Note that TOMT-GFP localizes to the cell body but not the stereocilia. Scale bar in (C) = 1 µm and applies to (C,D). Scale bar in (F) = 1 µm, and applies to (F,G).
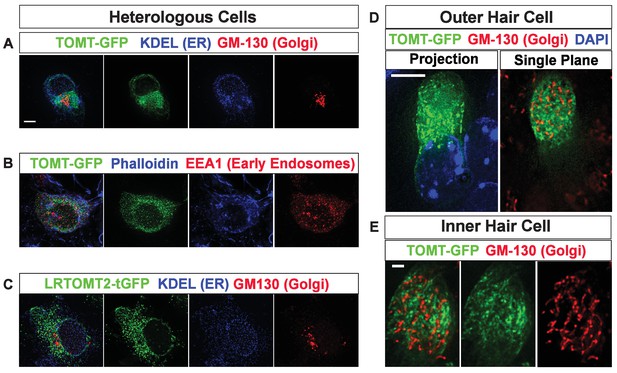
Subcellular localization of epitope-tagged TOMT constructs in heterologous cells and cochlear hair cells.
(A) HEK293 cells transfected with mTOMT-GFP (green), and immunostained for KDEL, a marker for the endoplasmic reticulum (ER, blue), and GM-130, a marker for the Golgi apparatus (Red). (B) HEK293 cells transfected with mTOMT-GFP (green), stained for phalloidin to label actin (blue), and immunostained with EEA1 to label early endosomes (red). (C) HEK293 cells transfected with human LRTOMT2-GFP (green) and immunostained with KDEL (blue) and GM130 (red). (D) An outer hair cell injectoporated at P3 with mTOMT-GFP (green), fixed 24 hr later, stained with DAPI to label nuclei (blue), and immunostained with GM130 (red). At left is a projection image showing mTOMT-GFP and DAPI labeling. At right is a single optical plane showing mTOMT-GFP and GM130 labeling. mTOMT-GFP was localized throughout the cytoplasm, without significant colocalization with GM130. (E) A single optical section from an inner hair cell injectoporated at P3 with mTOMT-GFP, fixed 24 hr later and immunostained with GM130 (red). Scale bar in (A) = 5 µm and applies to (A–C). Scale bar in (D) = 5 µm and (E) = 1 µm.
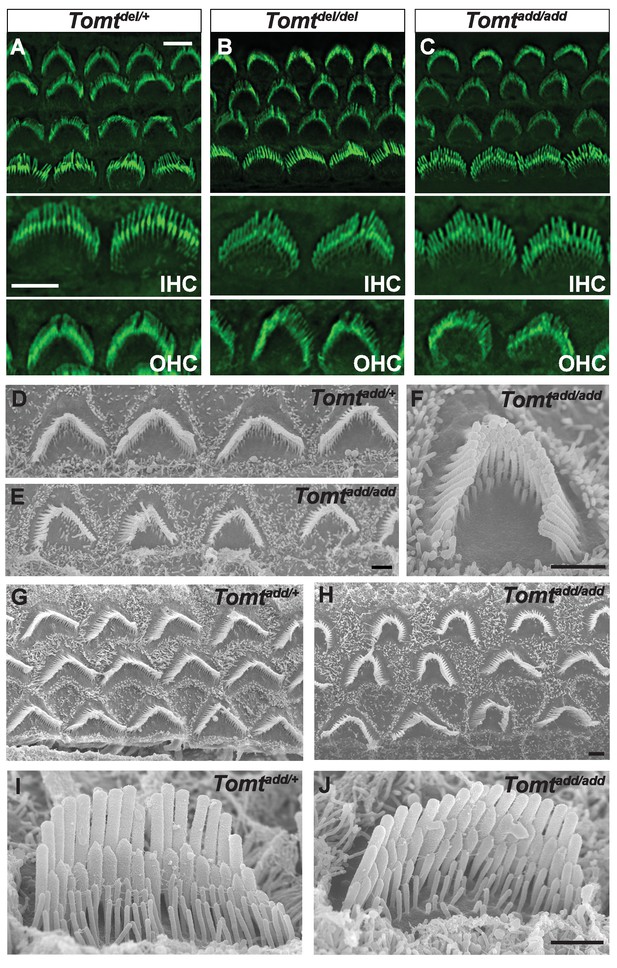
Morphology of hair bundles.
(A–C) P5 Cochlear whole mounts from the indicated mice stained with phalloidin to label stereocilia. Bottom panels show IHCs and OHCs from each genotype stained for phalloidin. Scale bar in (A) upper panel = 5 µm, applies to upper panels in (A–C). Scale bar in (A) lower panel = 5 µm, applies to lower panels in (A–C). (D–J) SEM analysis of cochlear hair bundles from P8 Tomtadd/+ and Tomtadd/add mice. (D) OHCs from middle region of P8 Tomtadd/+ cochlea. (E) OHCs from middle region of P8 Tomtadd/add cochlea. (F) Example of outer hair cell from middle region of P8 Tomtadd/add cochlea. (G) OHCs from base region of P8 Tomtadd/+ cochlea. (H) OHCs from base region of P8 Tomtadd/add cochlea. Note the presence of hair bundles with slightly more rounded phenotypes relative to P8 Tomtadd/+ cochlea. (I) IHC from middle region of P8 Tomtadd/+ cochlea. (J) IHC from middle region of P8 Tomtadd/add cochlea. Scale bar in (E) = 1 µm, applies to (D–E). Scale bar in (F) = 1 µm. Scale bar in (H) = 1 µm, applies to (G,H). Scale bar in (J) = 1 µm, applies to (I,J).
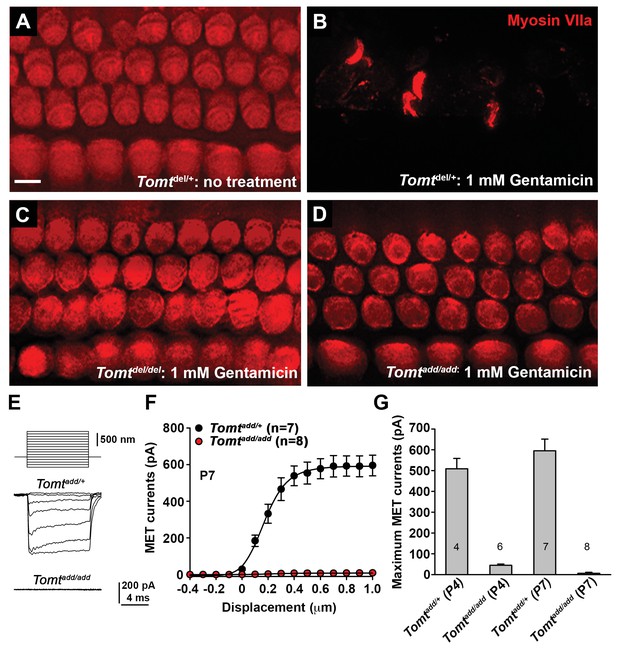
Analysis of mechanotransduction in Tomt mutants.
(A–D) P5 explants from middle region of the cochlea cultured for 24 hr with or without 1 mM Gentamicin, followed by fixation and immunostaining for MYO7A. (A) Tomtdel/+ cultured without Gentamicin. (B) Tomtdel/+ cultured for 24 hr with 1 mM Gentamicin. Note the almost complete loss of hair cells. (C) Tomtdel/del cultured for 24 hr with 1 mM Gentamicin. Hair cells are spared from Gentamicin-mediated ototoxicity. (D) Tomtadd/add cultured for 24 hr with 1 mM Gentamicin. Hair cells are spared from Gentamicin-mediated ototoxicity. (E) Representative mechanotransduction currents in OHCs from Tomtadd/+ and Tomtadd/add mice at P7 in response to a set of 10 ms hair bundle deflections ranging from −400 nm to 1000 nm (100 nm steps). (F) Current displacement plot obtained from similar data as shown in (E). Data are mean ± SEM. (G) Summary of data obtained from experiments as shown in (E–F) for Tomtadd/+ and Tomtadd/add at P4 and P7. Data are plotted as maximum MET currents. Number of cells tested is indicated on each column of the graph. Scale bar in (B) = 5 µm and applies to (A–D).
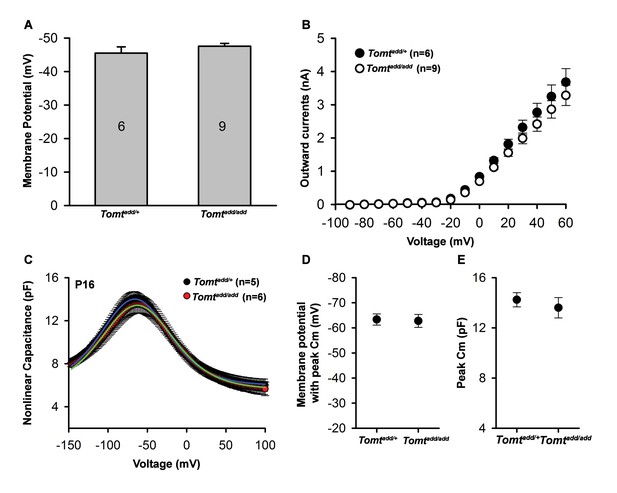
Basic membrane properties of cochlear hair cells in Tomt mutants.
(A) Membrane potential (in millivolts (mV)) of outer hair cells from P6 Tomtadd/+ and Tomtadd/add mice current-clamped at 0 picoamps. Number of cells is indicated on individual columns of the graph. (B) Outward currents (in nanoamps (nA)) elicited from outer hair cells of P6 Tomtadd/+ and Tomtadd/add mice at command potentials from −90 mV to 60 mV with a holding potential of −70 mV. (C). Nonlinear capacitance (in picofarads (pF)) as a function of voltage of outer hair cells from P16 Tomtadd/+ and Tomtadd/add mice. At right are summary data showing the membrane potential at which the peak Cm was recorded and the absolute value of the peak Cm, in pF. Data are mean ± SEM.
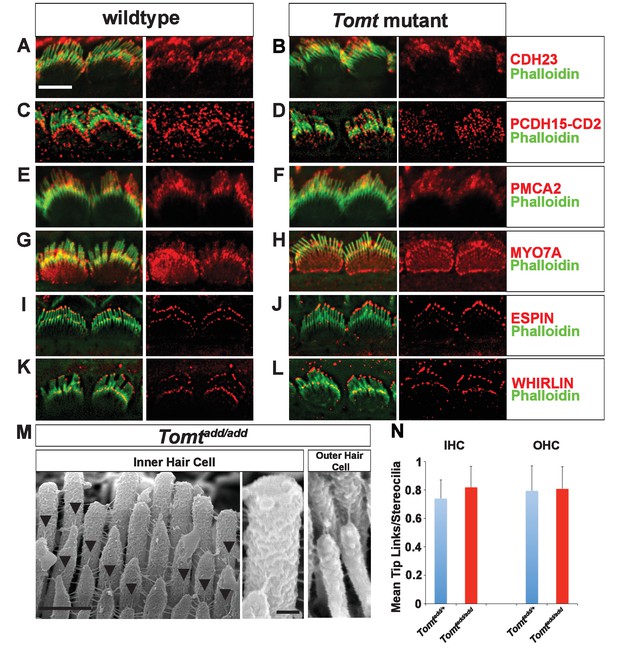
Analysis of tip links in Tomt mutants.
(A,B) P6 cochlear inner hair cells from (A) Tomtadd/+ and (B) Tomtadd/add mice stained with anti-Cdh23 (Siemens et al., 2004) and phalloidin. (C,D) P7 cochlear inner hair cells from (C) Tomtadd/+ and (D) Tomtadd/add mice stained with anti-PCDH15-CD2 (Webb et al., 2011) and phalloidin. (E,F) P6 cochlear inner hair cells from (E) Tomtadd/+ and (F) Tomtadd/add mice stained with anti-PMCA2 (Abcam) and phalloidin. (G,H) P8 cochlear inner hair cells from (G) Tomtadd/+ and (H) Tomtadd/add mice stained with anti-MYO7A (Proteus) and phalloidin. (I,J) P8 cochlear inner hair cells from (I) Tomtadd/+ and (J) Tom add/add mice stained with anti-Espin (BD Biosciences) and phalloidin. (K,L) P7 cochlear inner hair cells from (K) Tomtadd/+ and (L) Tomtadd/add mice stained with anti-Whirlin (see Materials and methods) and phalloidin. Scale bar in (A) = 5 µm, and applies to (A–L). (M) Tip links from middle region cochlear hair cells from P7-8 Tomtadd/add mice using SEM. Left and middle panels are from inner hair cell, right panel is from outer hair cell. Left panel scale bar = 500 nm, middle panel scale bar = 100 nm and applies to middle and right panels. (N) Quantification of tip-links per stereociliary column from P7-P8 middle region inner and outer hair cells from Tomtadd/+ (IHC n = 10 cells, OHC n = 9 cells) and Tomtadd/add (IHC n = 17 cells, OHC n = 9) mice.
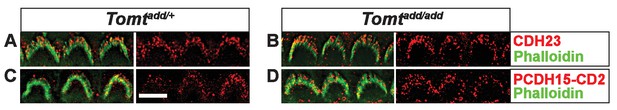
Immunostaining for tip-link components in outer hair cells from Tomt mutants.
(A,B) P6 cochlear outer hair cells from (A) Tomtadd/+ and (B) Tomtadd/add mice stained with anti-Cdh23 (Siemens et al., 2004) and phalloidin. (C,D) P7 cochlear inner hair cells from (C) Tomtadd/+ and (D) Tomtadd/add mice stained with anti-PCDH15-CD2 (Webb et al., 2011) and phalloidin. Scale bar in (A) = 5 µm and applies to (A–D).
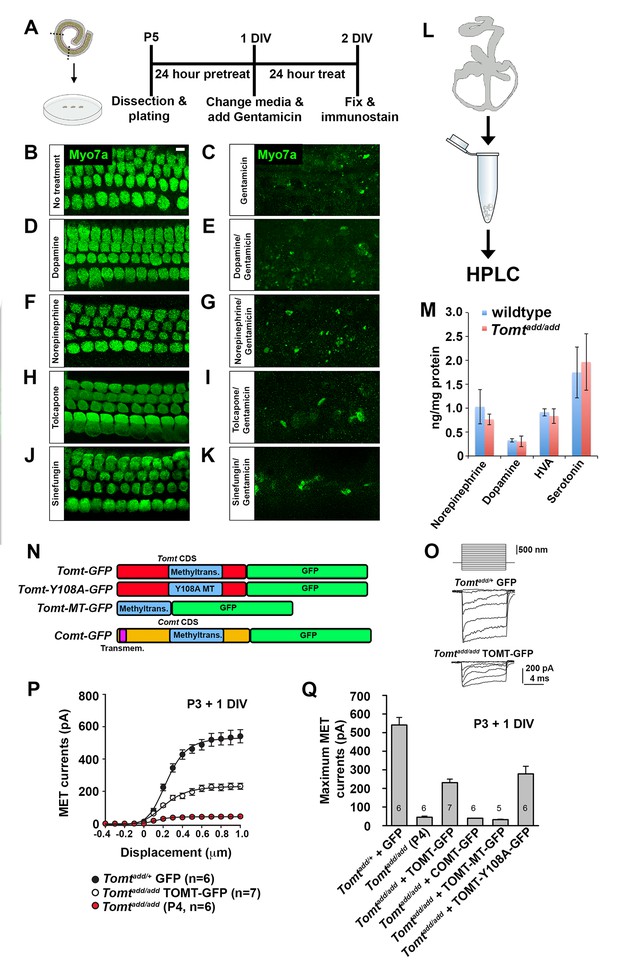
Effects of catecholamines on mechanotransduction.
(A) Experimental paradigm for assaying the effect of catecholamines on Gentamicin-mediated ototoxicity. The organ of Corti of P5 C57BL/6J mice was dissected, sectioned into three parts, and plated for in vitro culture. Explants were pretreated with catecholamines or inhibitors for 24 hr. Media was replaced with fresh media containing catecholamines with or without 1 mM Gentamicin. Explants were cultured for 24 additional hours, followed by fixation and immunostaining for MYO7A (green) to reveal hair cells. (B) Untreated P5 explants cultured without catecholamine or Gentamicin. (C) P5 explants cultured with Gentamicin. Note the significant loss of hair cells. (D) P5 explants treated for 48 hr with 2.5 µM Dopamine, with a media change with fresh Dopamine at 24 hr. (E) P5 explants pretreated for 24 hr with 2.5 µM Dopamine, followed by 24 hr of 2.5 µM Dopamine and 1 mM Gentamicin. Dopamine treatment does not rescue cells from Gentamicin-mediated ototoxicity. (F) P5 explants treated for 48 hr with 1 µM Norepinephrine, with a media change with fresh Norepinephrine at 24 hr. (G) P5 explants pretreated for 24 hr with 1 µM Norepinephrine, followed by 24 hr of 1 µM Norepinephrine and 1 mM Gentamicin. Norepinephrine treatment does not rescue cells from Gentamicin-mediated ototoxicity. (H) P5 explants treated for 48 hr with 10 µM Tolcapone, a COMT inhibitor, with a media change with fresh Tolcapone at 24 hr. (I) P5 explants pretreated for 24 hr with 10 µM Tolcapone, followed by 24 hr of 10 µM Tolcapone and 1 mM Gentamicin. Tolcapone treatment does not rescue cells from Gentamicin-mediated ototoxicity. (J) P5 explants treated for 48 hr with 1 µM Sinefungin, a global methyltransferase inhibitor, with a media change with fresh Sinefungin at 24 hr. (K) P5 explants pretreated for 24 hr with 1 µM Sinefungin, followed by 24 hr of 1 µM Sinefungin and 1 mM Gentamicin. Sinefungin treatment does not rescue cells from Gentamicin-mediated ototoxicity. Scale bar in (B) = 5 µm, applies to (B–K). (L) Experimental paradigm for assaying catecholamine levels of inner ears from P5 wildtype (C57BL/6J) and Tomtadd/add using HPLC. Inner ears from P5 wildtype and Tomtadd/add mice were dissected and snap frozen on dry ice. Each sample contained two temporal bones pooled together and represented one animal. Catecholamine levels were assayed using HPLC and normalized to total protein content. (M) Quantification of catecholamine levels. Norepinephrine, Adrenaline, DOPAC, Dopamine, 5-HIAA, HVA, Serotonin, and 3-MT were assayed, but only Norepinephrine, Dopamine, HVA, and Serotonin were detected. Absolute levels were normalized to total protein. Values represent mean ± SD of C57BL/6 (n = 5 animals, 10 temporal bones) or Tomtadd/add (n = 5 animals, 10 temporal bones). (N) Schematic for constructs used for injectoporation experiments. Constructs included mouse TOMT coding sequence (CDS) tagged at the C terminus with GFP (Tomt-GFP), mouse TOMT CDS with Y108A mutation (orthologous conserved amino acid critical for COMT enzymatic activity; [Zhang and Klinman, 2011]) tagged at the C-terminus with GFP (Tomt-Y108A-GFP), mouse TOMT methyltransferase (MT) domain (based on NCBI conserved protein domain family cl17173) tagged at the C-terminus with GFP (Tomt-MT-GFP), and mouse COMT CDS tagged at the C-terminus with GFP (Comt-GFP). Methyltransferase and transmembrane domains indicated. (O) Representative mechanotransduction currents in P3+1 DIV OHCs from Tomtadd/+ injectoporated at P3 with GFP and Tomtadd/add injectoporated at P3 with TOMT-GFP in response to a set of 10 ms hair bundle deflections ranging from −400 nm to 1000 nm (100 nm steps). (P) Current displacement plots obtained from similar data as shown in O. (Q) Plot of maximum MET currents obtained from similar data as in (O–P) from OHCs injectoporated with constructs described in (N). Data are mean ± SEM.
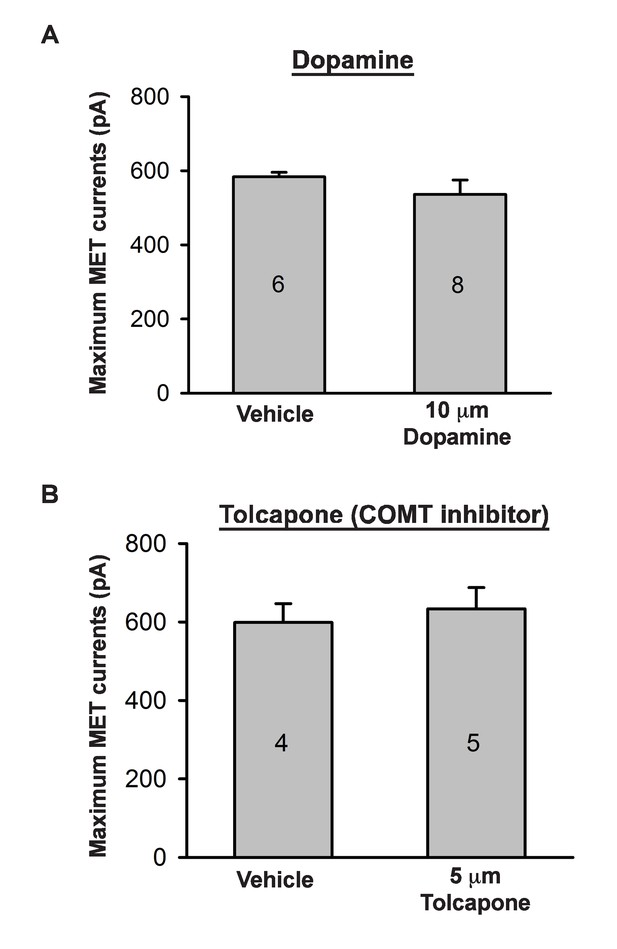
Effects of catecholamine manipulation on mechanotransduction.
(A) Plot of maximum mechanoelectrical transduction currents evoked in outer hair cells from P6 cochlear explants pretreated overnight with vehicle or 10 µM Dopamine in response to 10 ms hair bundle deflections ranging from −400 nm to 1000 nm (100 nm steps). Numbers on columns indicate number of cells tested. (B) Plot of maximum mechanoelectrical transduction currents evoked in P6 outer hair cells from cochlear explants treated with vehicle or 5 µM Tolcapone in the recording pipette in response to 10 ms hair bundle deflections ranging from −400 nm to 1000 nm (100 nm steps). Numbers on columns indicate number of cells tested.
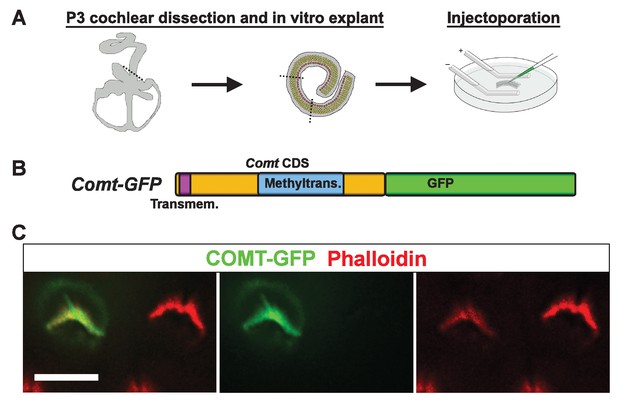
mCOMT-GFP expression in cochlear hair cells.
(A) Depiction of experimental paradigm for injectoporation of epitope-tagged constructs (as [Xiong et al., 2014]). The organ of Corti of P3 C57BL/6J mice was dissected, sectioned into three parts, and plated as in vitro explants. Plasmid DNA was microinjected into the space surrounding the basal compartment of hair cells, followed by electroporation. Injectoporated explants were cultured for 24 hr, fixed, and immunostained. (B) Schematic depicting construct used for injectoporation. Mouse COMT coding sequence (CDS) was tagged at the C-terminus with GFP (Comt-GFP). Transmembrane and methyltransferase domains are indicated. (C) OHCs from P3 C57BL/6J mice were injectoporated with mCOMT-GFP, and stained for phalloidin to label actin. mCOMT-GFP localizes to the hair bundle and kinocilium. Scale bar = 5 µm.
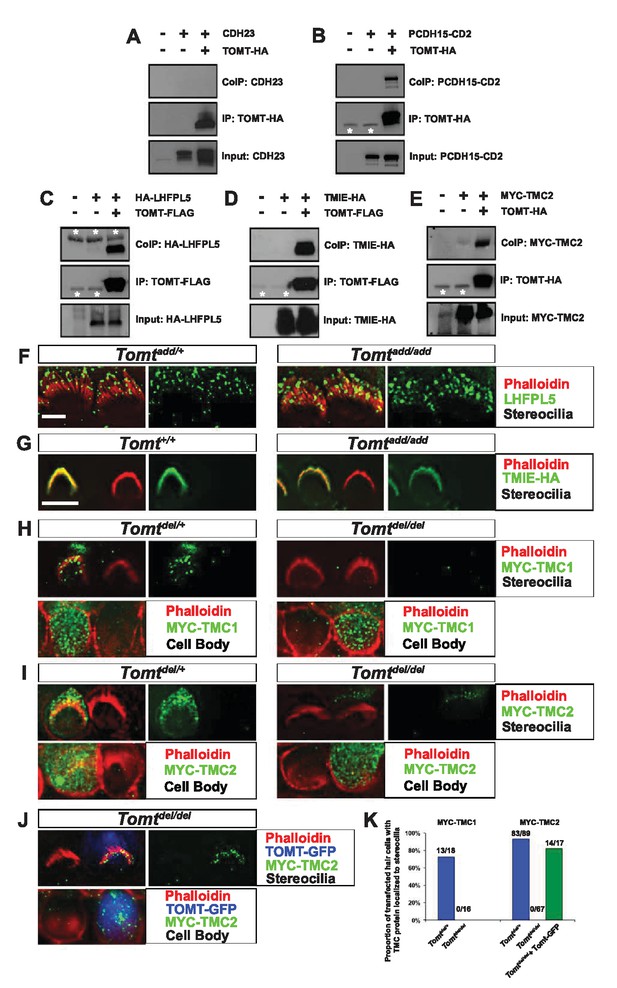
Protein-protein interactions with mTOMT.
(A–E) HEK293 cells were transfected with the constructs indicated on top of each panel to perform co-immunoprecipitation (CoIP) experiments. Immunoprecipitations were carried out with HA (A,B,E) or Flag (C,D) antibodies, followed by Western blotting to detect proteins. The upper panels show CoIP results, the middle row shows IP results and the lower rows show input protein. White asterisks indicate 25 kDa light-chain IgG bands from antibodies used for IP. (F) P6 whole mount cochleas from Tomtadd/+ (left panels) and Tomtadd/add mice (right panels) stained with anti-TMHS/LHFPL5 (Xiong et al., 2012) and phalloidin. Scale bar in (F) = 5 µm, applies to both panels. (G) OHCs from P3 wild-type (C57BL/6J) (left) and Tomtadd/add (right) mice were injectoporated with TMIE-HA, cultured for 24 hr, fixed and stained with anti-HA antibody and phalloidin. TMIE-HA localizes to the hair bundle of both wildtype and Tomtadd/add OHCs. (H) Outer hair cells from P3 Tomtdel/+ (left) and Tomtdel/del (right) mice were injectoporated with MYC-TMC1, cultured for 24 hr, fixed and stained with anti-MYC antibody and phalloidin. MYC-TMC1 localizes to the hair bundle of Tomtdel/+ but is absent from the hair bundle of Tomtdel/del OHCs. Optical sections at the level of the hair bundle and cell body are shown. (I) Outer hair cells from P3 Tomtdel/+ (left) and Tomtdel/del (right) mice were injectoporated with MYC-TMC2, cultured for 24 hr, fixed and stained with anti-MYC antibody and phalloidin. MYC-TMC2 localizes to the hair bundle of Tomtdel/+ but is absent from the hair bundle of Tomtdel/del OHCs. Optical sections at the level of the hair bundle and cell body are shown. (J) Outer hair cells from P3 Tomtdel/del mice were injectoporated with mTOMT-GFP and MYC-TMC2, cultured for 24 hr, fixed and stained with anti-MYC antibody and phalloidin. MYC-TMC2 localizes to the hair bundle of Tomt del/del hair cells in the presence of exogenous mTOMT-GFP. Optical sections at the level of the hair bundle and cell body are shown. (K) Quantification of injectoporation experiments shown in (H–J). The data are plotted as the proportion of injectoporated outer hair cells for each genotype and construct that exhibited TMC protein localized to stereocilia. The proportion of cells for each condition are indicated on the columns. Only healthy transfected hair cells, as determined by stereocilia morphology, were included for analysis. Scale bar in (G) = 5 µm, applies to (G–J).
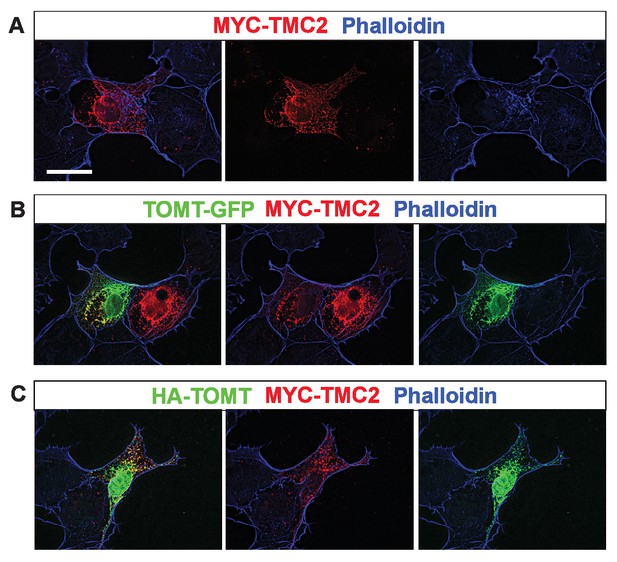
Analysis of protein localization in heterologous cells.
(A–C) COS-7 cells were transfected with MYC-TMC2 alone (A); mTOMT-GFP and MYC-TMC2 (B); or HA-mTOMT and MYC-TMC2 (C). After 24 hr, cells were fixed and stained for phalloidin (blue), anti-MYC (red), and anti-HA (green). Scale bar in left column = 25 µm and applies to all images.
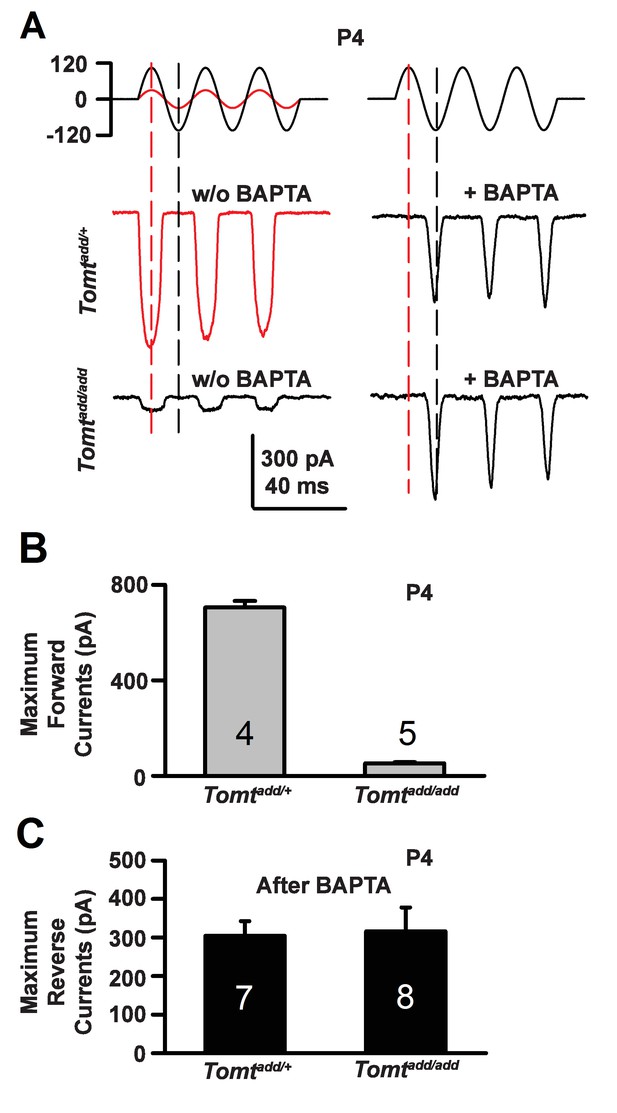
Forward and reverse polarity currents in Tomt mutants.
(A) Representative mechanotransduction currents in response to sinusoidal deflection of hair bundles at P4 for OHCs from Tomtadd/+ and Tomtadd/add mice with and without 5 mM BAPTA treatment. All recordings were from middle or apical OHCs at a holding potential of −70 mV. The stimulus monitor (the driving voltage to the fluid jet) is shown at the top. A lower voltage (red trace) was necessary to obtain similar displacements in Tomtadd/+ mice relative to Tomtadd/add mice (black trace). Positive driving voltage denotes displacement toward the tallest edge of the hair bundle. (B) Plot of maximum forward MET currents obtained from similar data as A. Number of cells for each condition is indicated on graph. (C) Plot of maximum reverse currents obtained from similar data as in (A). Number of cells for each condition is indicated on graph.





