PTEN controls glandular morphogenesis through a juxtamembrane β-Arrestin1/ARHGAP21 scaffolding complex
Figures
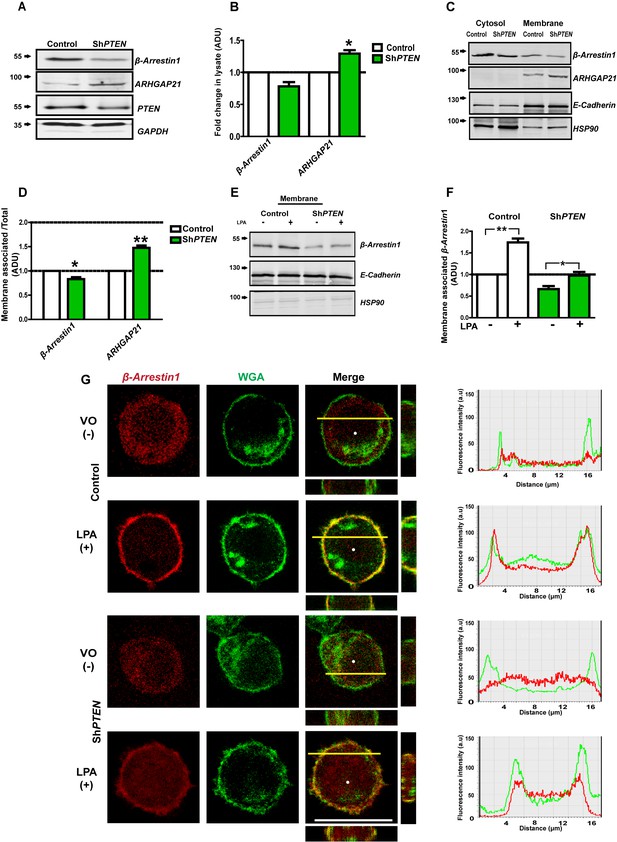
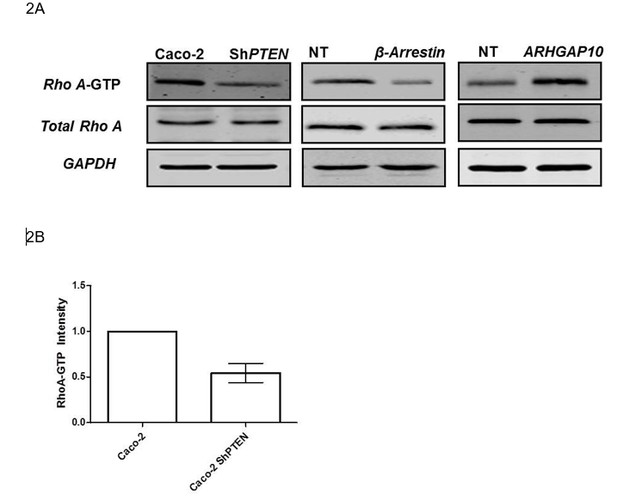
PTEN enhances membrane recruitment of β-Arrestin1.
(A,B) Total lysate β-Arrestin1 and ARHGAP21 expression in ShPTEN vs Caco-2 (control). GAPDH loading control; β-Arrestin1 = 0.78 ± 0.06; p=NS (0.08); ARHGAP21 = 1.29 ± 0.05;*p=0.03. (C) Cytosol and membrane localization of β-Arrestin1 and ARHGAP21 in control Caco-2 or Caco-2 ShPTEN (ShPTEN) cells. E-Cadherin and HSP90 used as cell membrane or cytosol markers. (D) Summary β-Arrestin1 and ARHGAP21 membrane ADU in ShPTEN cells vs Caco-2 control. Membrane values are normalized against total lysate ADU values for each protein (β-Arrestin1 = 0.82 ± 0.03 *p<0.05; ARHGAP21 = 1.48 ± 0.05 **p<0.01). (E) β-Arrestin1 expression in membrane fractions of control Caco-2 and ShPTEN cells after treatment. (F) Summary fold change of membrane β-Arrestin1 after LPA (+) or VO [vehicle only(-)] treatment shown in (E) ;Caco-2 (+) vs (-)=1.74 ± 0.09; **p<0.01; ShPTEN (+) vs ShPTEN (-)=0.97 ± 0.08 vs 0.66 ± 0.07;*p<0.05 values expressed as fold change relative to Caco-2 (-) control. ShPTEN (-) vs Caco-2 (-)=0.66 ± 0.07;*p<0.05; ShPTEN (+) vs Caco-2 (+)=0.97 ± 0.08 vs 1.74 ± 0.09;**p<0.01. Green bars indicate ShPTEN cells. (G) Plasma membrane localization of the β-Arrestin1-mCherry fusion protein in Caco-2 control (top two panels) or ShPTEN (bottom two panels) after VO (-) or LPA (+) treatment. Red and green fluorescence emitted by m-Cherry and Alexa 488 labels correspond to β-Arrestin1 and WGA, respectively. Note WGA localization to plasma membranes and Golgi apparatus. Colocalization of β-Arrestin1 and WGA at the plasma membrane revealed by composite yellow signal in Merge, Z-stack images and by overlap of fluorescence intensity curves in line scans. Horizontal yellow bars indicate focal plane. All experiments in triplicate. Analyses by Student’s paired t test or ANOVA with Tukey post hoc test. White scale bar 20 µm. Molecular weights indicated by arrows in blots.
-
Figure 1—source data 1
Source data for Figure 1B.
- https://doi.org/10.7554/eLife.24578.010
-
Figure 1—source data 2
Source data for Figure 1D.
- https://doi.org/10.7554/eLife.24578.011
-
Figure 1—source data 3
Source data for Figure 1F.
- https://doi.org/10.7554/eLife.24578.012
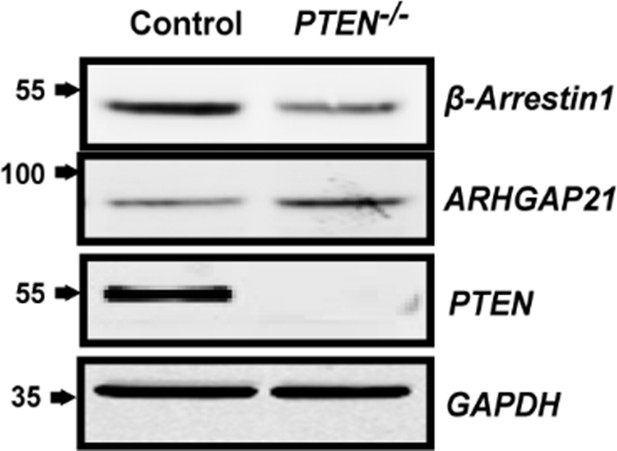
Total β-Arrestin1 and ARHGAP21 expression in control HCT116 and PTEN -/- cell lysates.
GAPDH loading control.
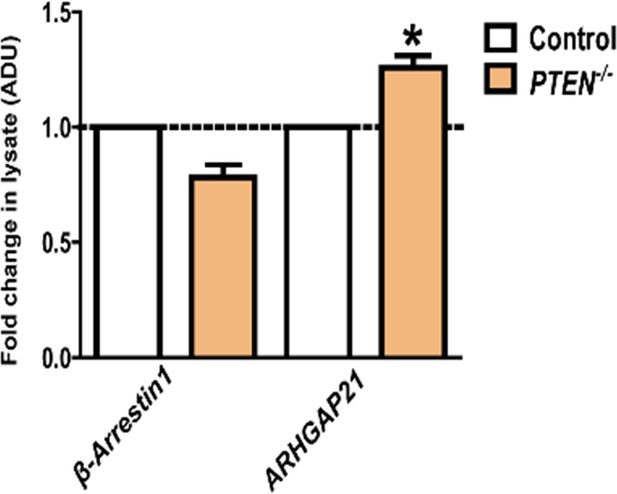
Cell lysate expression of β-Arrestin1 and ARHGAP21 shown in Figure 1—figure supplement 1.
Values in PTEN -/- cells are expressed as fold change relative to HCT116 control - β-Arrestin1 = 0.78 ± 0.06; p=NS (0.06); ARHGAP21 = 1.53 ± 0.09; *p=0.03. Orange bars indicate PTEN -/- cells.
-
Figure 1—figure supplement 2—source data 1
. Figure 1—figure supplement 2 Beta-Arestin1 and ARHGAP21 expression in HCT116 clones -Source data.
- https://doi.org/10.7554/eLife.24578.013
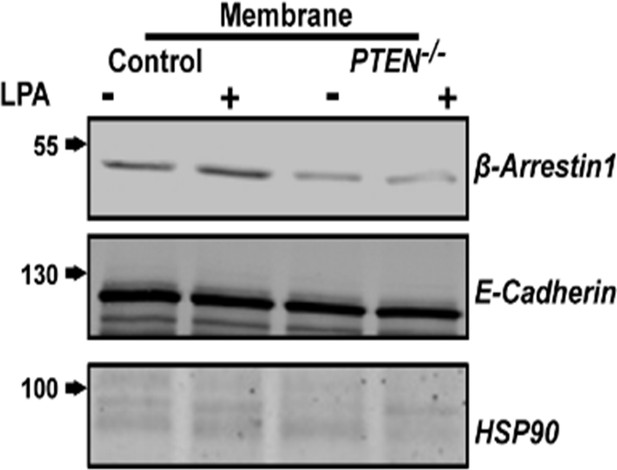
Treatment effects on membrane β-Arrestin1 in HCT116 control vs PTEN -/- cells.
E-Cadherin and HSP90 used as membrane and cytosolic markers, respectively.
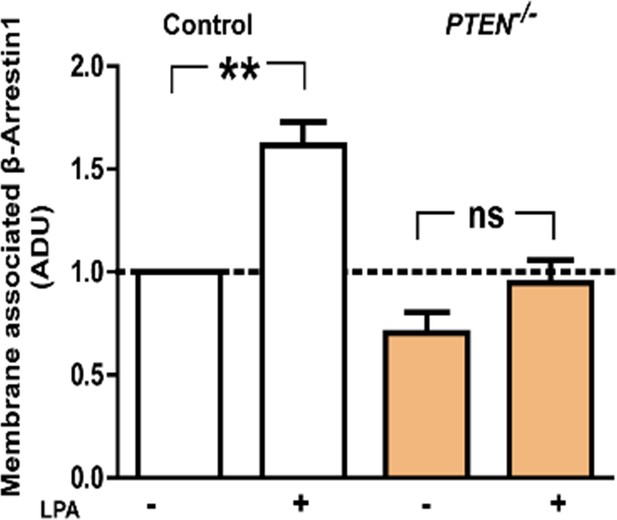
Treatment effects on membrane β-Arrestin1 shown in Figure 1—figure supplement 3.
Fold change of membrane β-Arrestin1 after LPA (+) or VO [vehicle only (-)] treatment; HCT116 control cells (+) vs (-)=1.62 ± 0.11; **p=0.01; PTEN -/- (+) vs PTEN -/- (-)=0.94 ± 0.11 vs 0.70 ± 0.10; p=NS; HCT116 (-) vs PTEN -/- (-); p=NS. Values are expressed as fold change relative to HCT116 (-) control. Membrane values were normalized against total lysate ADU values for each protein.
-
Figure 1—figure supplement 4—source data 1
Figure 1—figure supplement 4 LPA effects on Beta-Arrestin1 in HCT116 clones.
- https://doi.org/10.7554/eLife.24578.014
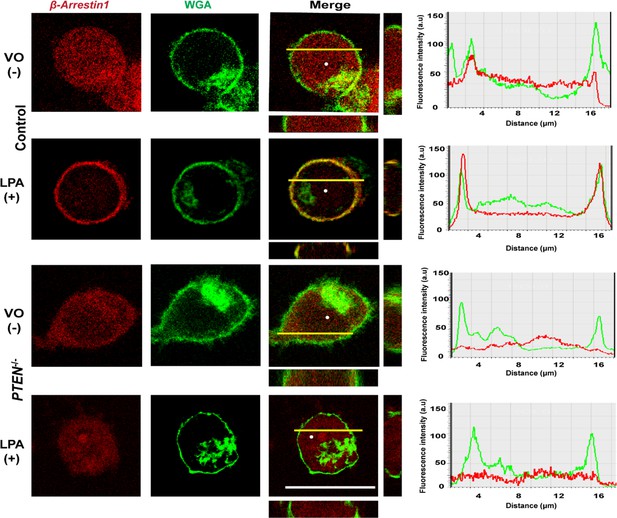
Plasma membrane localization of the β-Arrestin1-mCherry fusion protein in HCT116 control (top two panels) or PTEN -/- cells (bottom two panels) after VO (-) or LPA (+) treatment.
Red and green fluorescence emitted by m-Cherry and Alexa 488 labels correspond to β-Arrestin1 and WGA signals. Colocalization revealed by composite yellow signal in Merge, Z-stack images and overlap of fluorescence intensity curves in line scans. Data analysis by ANOVA or Student’s paired t test. Horizontal yellow bars indicate focal plane.
Control mCherry distribution in HCT116 and PTEN -/- cells.
Scale bars (white) 20 µm. Molecular weights indicated by arrows in blots.
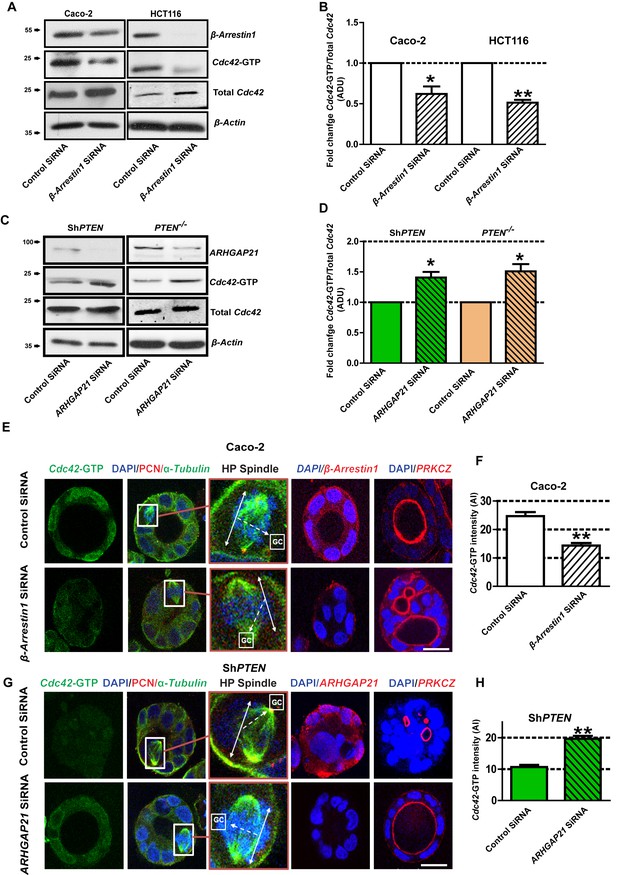
PTEN controls morphogenesis through Cdc42/β-Arrestin1/ARHGAP21 interactions.
(A,B) SiRNA β-Arrestin1 knockdown (KD) suppresses Cdc42-GTP in Caco-2 and HCT116 cells (fold changes = 0.62 ± 0.09;*p=0.03 and 0.51 ± 0.03;**p=0.005 respectively. (C,D) SiRNA ARHGAP21 KD enhances Cdc42-GTP in Caco-2 ShPTEN (ShPTEN) and HCT116 PTEN -/- (PTEN -/-) cells (fold change = 1.41 ± 0.09 and 1.51 ± 0.11, respectively; *p=0.02 for each. Cdc42-GTP ADU was normalized against total Cdc42. (E) SiRNA β-Arrestin1 KD suppresses Cdc42-GTP signal intensity, impairs spindle orientation and inhibits single lumen formation. High-power (HP) spindle views (orange border) enlarge areas within white rectangles and show orientation angles (interrupted white arrows) of spindle planes (double-headed solid white arrows) toward gland centres (GCs). Normal spindle planes are orientated at approximately 900 angles relative to gland centres [GCs] (Jaffe et al., 2008). Summary SiRNA effects on spindle angles relative to GCs are shown in Figure 2—figure supplement 1. (F) Summary SiRNA effects on Cdc42-GTP intensity shown in (E) - control vs β-Arrestin1 SiRNA = 24.67 ± 1.45 vs 14.33 ± 0.88 AI units; **p=0.004. β-Arrestin1 KD also suppresses single central lumen formation in 3D Caco-2 cultures (E; Figure 2—figure supplement 2). (G) SiRNA ARHGAP21 KD increases Cdc42-GTP signal intensity (H), rescues spindle orientation (G, Figure 2—figure supplement 3) and central lumen formation in ShPTEN 3D cultures (G, Figure 2—figure supplement 4). (H) Cdc42-GTP, control vs SiRNA ARHGAP21 KD in ShPTEN cultures = 10.67 ± 0.67 vs=19.67 ± 0.88 AI units; **p<0.01. Assays at 4 days of culture. Imaging Cdc42-GTP [green], pericentrin (PCN) [red], α-Tubulin [green], ARHGAP21 [red], PRKCZ [red], β-Arrestin1 [red] and DAPI [blue]. All experiments conducted in triplicate. All analyses by paired Student’s t test. Scale bars 20 µm. Molecular weights indicated by arrows in blots.
-
Figure 2—source data 1
Source data for Figure 2B.
- https://doi.org/10.7554/eLife.24578.020
-
Figure 2—source data 2
Source data for Figure 2D.
- https://doi.org/10.7554/eLife.24578.021
-
Figure 2—source data 3
Source data for Figure 2F.
- https://doi.org/10.7554/eLife.24578.022
-
Figure 2—source data 4
Source data for Figure 2H.
- https://doi.org/10.7554/eLife.24578.023
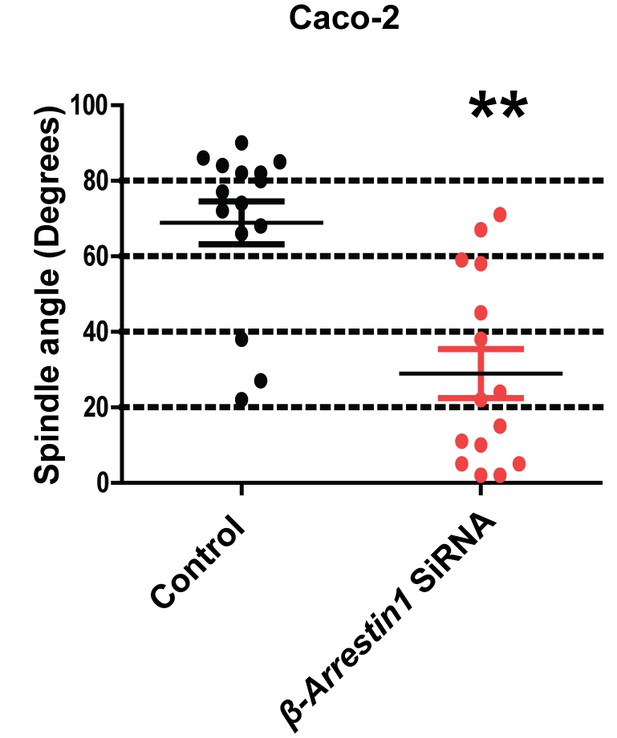
Summary effects of siRNA β-Arrestin1 or ARHGAP21 KD vs control non-targeting SiRNA on mitotic spindle angles and lumen formation in 3D Caco-2 and Caco-2 ShPTEN (ShPTEN) cultures.
Effects of control SiRNA vs β-Arrestin1 SiRNA on spindle angles (68.9 ± 5.600 vs 28.9 ± 6.490; **p<0.01) in 3D Caco-2 cultures. Green bars indicate ShPTEN cultures. All analyses by paired Student’s t test.
-
Figure 2—figure supplement 1—source data 1
Figure 2—figure supplement 1 Spindle angles in Caco-2 after Beta-Arrestin1 KD.
- https://doi.org/10.7554/eLife.24578.024
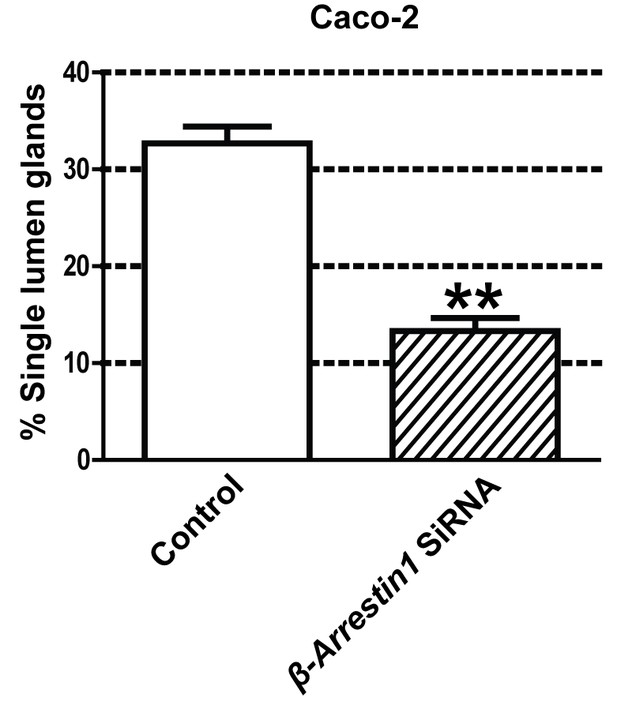
Summary effects of siRNA β-Arrestin1 or ARHGAP21 KD vs control non-targeting SiRNA on mitotic spindle angles and lumen formation in 3D Caco-2 and Caco-2 ShPTEN (ShPTEN) cultures.
Effects of control SiRNA vs β-Arrestin1 SiRNA on percentage single lumen formation (32.67 ± 1.76% vs 12.67 ± 1.76%;**p<0.01) in 3D Caco-2 cultures. Green bars indicate ShPTEN cultures. All analyses by paired Student’s t test.
-
Figure 2—figure supplement 2—source data 1
Figure 2—figure supplement 2 - Single central lumen fomation on Caco-2 after Beta-Arrestin1 KD.
- https://doi.org/10.7554/eLife.24578.025
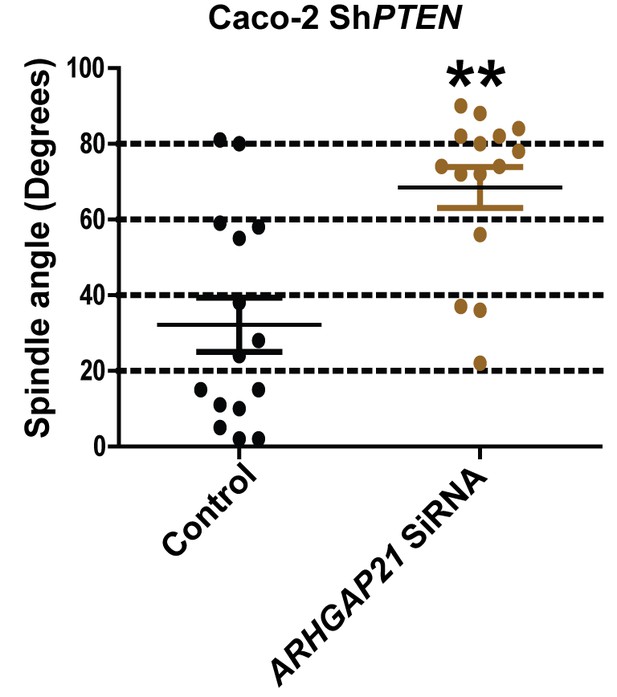
Summary effects of siRNA β-Arrestin1 or ARHGAP21 KD vs control non-targeting SiRNA on mitotic spindle angles and lumen formation in 3D Caco-2 and Caco-2 ShPTEN (ShPTEN) cultures.
Effects of control SiRNA vs ARHGAP21 SiRNA on spindle angles (32.2 ± 7.17 vs 68.5 ± 5.400’; **p<0.01), in 3D Caco-2 ShPTEN (ShPTEN) cultures. Green bars indicate ShPTEN cultures. All analyses by paired Student’s t test.
-
Figure 2—figure supplement 3—source data 1
Figure 2—figure supplement 3 Spindle angles in ShPTEN after ARHGAP21 KD.
- https://doi.org/10.7554/eLife.24578.026
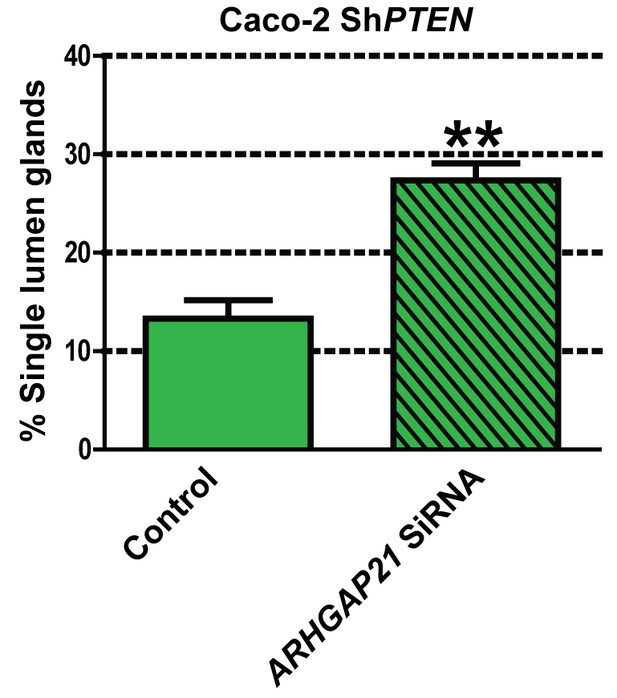
Summary effects of siRNA β-Arrestin1 or ARHGAP21 KD vs control non-targeting SiRNA on mitotic spindle angles and lumen formation in 3D Caco-2 and Caco-2 ShPTEN (ShPTEN) cultures.
Effects of control SiRNA vs ARHGAP21 SiRNA on percentage single lumen formation (13.33 ± 1.33% vs 27.33 ± 1.76%; **p=0.01) in 3D Caco-2 ShPTEN (ShPTEN) cultures. Green bars indicate ShPTEN cultures. All analyses by paired Student’s t test.
-
Figure 2—figure supplement 4—source data 1
Figure 2—figure supplement 4 Single central lumen in ShPTEN after ARHGAP21kd.
- https://doi.org/10.7554/eLife.24578.027
PTEN C2 enhances β-Arrestin1-ARHGAP21 binding.
(A) β-Arrestin1-ARHGAP21 CoIPs in PTEN-expressing and -deficient cells. β-Arrestin1-associated ARHGAP21 shown in top panel against a constant β-Arrestin1 bait signal (second panel). IgG-negative controls. Total β-Arrestin1 and ARHGAP21 in lysates and GAPDH loading controls shown in lower three panels. (B) Summary β-Arrestin1-associated ARHGAP21 in PTEN-deficient (colored bars) vs PTEN-expressing cells (clear bars). Values normalized against total ARHGAP21 = 0.35 ± 0.01;**p<0.01 (ShPTEN) and 0.39 ± 0.02;**p<0.01 (PTEN -/ - cells), respectively. (C) Schematic of GFP-labeled PTEN constructs used (top to bottom - wild type (wt) PTEN; PTEN - MCBR3 membrane binding mutant; catalytically inactive PTEN C124S; PTEN C124S - A4 (CS-A4) and PTEN C124S -T383A (CS-T383A) mutants that lack or retain β-Arrestin1 binding capacity, respectively (Lima-Fernandes et al., 2011), C2 and the C2-MCBR3 membrane binding mutant. (D) β-Arrestin1-associated ARHGAP21 in ShPTEN cells after transfection with GFP-labeled-EV control vs- C2 or -C2-MCBR3 (top panel). β-Arrestin1 bait signal shown in second panel. Total β-Arrestin1 and ARHGAP21 in lysates shown in third and fourth panels. Expression of GFP-labeled C2, C2-MCBR3 and EV and GAPDH loading controls shown in two lowest panels. (E) Summary fold change of β-Arrestin1-associated ARHGAP21 vs EV control; C2 = 2.51 ± 0.08;**p<0.01 or C2-MBCR3 = 1.03 ± 0.06; p=NS. β-Arrestin1-associated ARHGAP21 normalized against total ARHGAP21 in lysate. (F) Proximity ligation assay (PLA) of β-Arrestin1 interactions with PTEN constructs (red fluorescence) in PTEN -/- cells. Top row - GFP-labeled CS-T383A, CS-A4, C2, C2-MCBR3; Bottom row - positive control - HCT116 cells; negative control - PTEN -/- cells transfected with GFP only. (G) β-Arrestin1-ARHGAP21 interactions. Top row PTEN -/- cells transfected with GFP-labeled full-length PTEN, -PTEN-MCBR3, -C2 and -C2-MCBR3; Bottom row - positive control - HCT116 cells; negative control - PTEN -/- cells transfected with GFP only. Scale bars - 20 µm; Molecular weights indicated by arrows in blots.
-
Figure 3—source data 1
Source data for Figure 3B.
- https://doi.org/10.7554/eLife.24578.034
-
Figure 3—source data 2
Source data for Figure 3E.
- https://doi.org/10.7554/eLife.24578.035
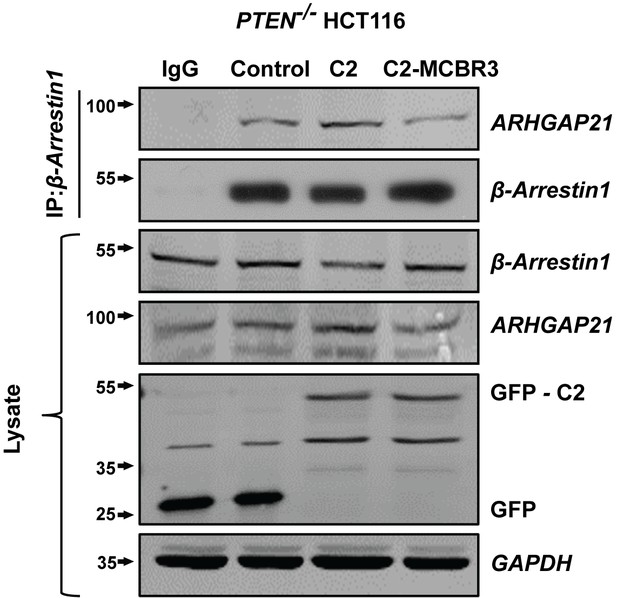
Top panel - Effects of GFP-labelled empty vector (EV) control, C2 and C2-MCBR3-GFP on β-Arrestin1-associated ARHGAP21 in PTEN -/- cells.
Second panel - β-Arrestin1 bait signal. IgG-negative controls shown. Panels 3–6 Total β-Arrestin1, total ARHGAP21, expression of GFP-labeled C2, C2-MCBR3 or EV control constructs and GAPDH loading controls.
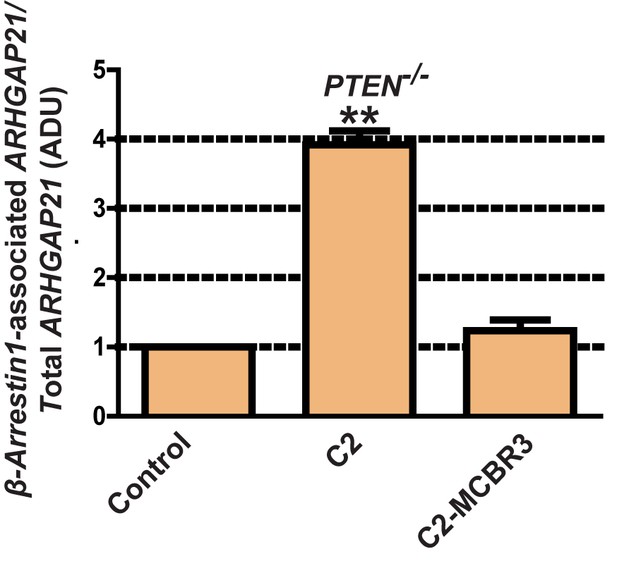
Fold changes of β-Arrestin1-associated ARHGAP21 shown in Figure 3—figure supplement 1 vs EV control.
C2 = 3.92 ± 0.2;**p<0.01; C2-MCBR3 = 1.23 ± 0.15 ADU; C2-MCBR3 vs EV, p=NS. β-Arrestin1-associated ARHGAP21 normalized against total ARHGAP21.
-
Figure 3—figure supplement 2—source data 1
Figure 3—figure supplement 2 Transfection effects on Beta-Arrestin1-associated ARHGAP21
- https://doi.org/10.7554/eLife.24578.036
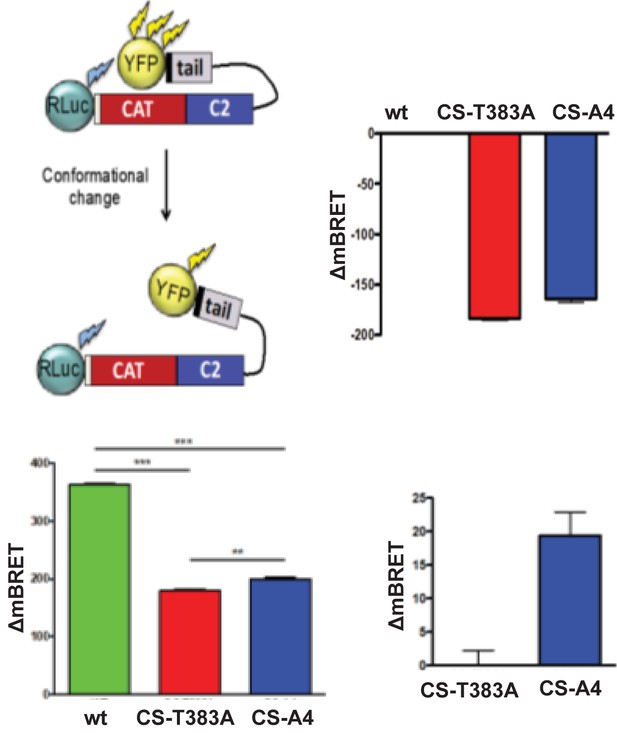
Diagram of Rluc-PTEN-YFP illustrating how conformational changes may alter BRET measurements, although the real orientations of donor and acceptor proteins are not known.
mBRET measurements in HEK cells transfected with Rluc-PTEN-YFP (wt), Rluc-PTEN CS - T383A-YFP or PTEN CS - A4-YFP. Data shown represent mean ± sem for four independent experiments and indicate different conformations between wt PTEN, CS - T383A and CS - A4.
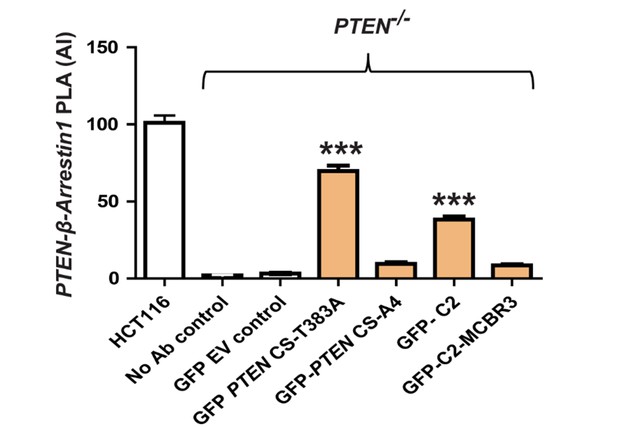
PTEN:β-Arrestin1 interaction AI after transfection of PTEN -/- cells by CS-T383A, CS-A4, C2 or C2-MCBR3.
Vs EV control shown in (F); CS-T383A = 69.83 ± 3.67; CS-A4 = 9.62 ± 0.99; C2 = 38.3 ± 2.09; C2-MCBR3 = 8.66 ± 0.81 AI units;**p<0.01. EV control vs CSTA4 or C2-MCBR3 = NS. Positive control - PTEN:β-Arrestin1 interactions in HCT116 cells = 101 ± 4.9; negative controls - No antibody (2.12 ± 0.25 and PTEN -/- cells transfected with GFP-EV only = 3.21 ± 0.48).
-
Figure 3—figure supplement 4—source data 1
Figure 3—figure supplement 4 PLA analysis of PTEN:Beta-Arrestin1 interactions.
- https://doi.org/10.7554/eLife.24578.037
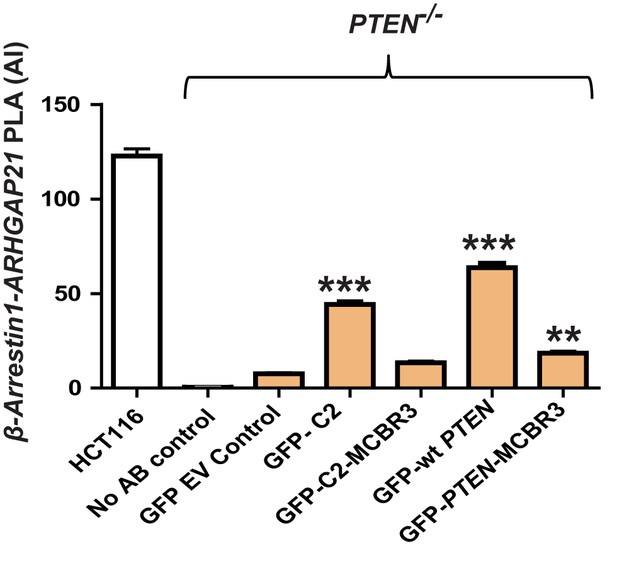
β-Arrestin1-ARHGAP21 interaction AI after transfection of PTEN -/- cells by GFP-labelled C2, C2-MCBR3, wt PTEN or PTEN-MCBR3.
C2 = 44.23 ± 1.92; C2-MCBR3 = 13.47 ± 0.88; wt PTEN = 63.7 ± 2.35; PTEN M-CBR3 = 18.53 ± 0.90 AI units; Positive control - β-Arrestin1:ARHGA21 interactions in HCT116 cells = 122.8 ± 3.8; negative controls - No antibody (0.39 ± 0.09 and PTEN -/- cells transfected with GFP-EV only = 7.58 ± 0.56). ***p<0.001; For C2 and wt PTEN vs EV (***) p<0.001; For PTEN-MCBR3 vs EV; (**) - p<0.01; For EV vs C2-MCBR3 and C2-MCBR3 vs PTEN-MCBR3, p=NS (n = 35 cells for each PLA experimental condition in triplicate). Red, blue and green bars - BRET assays. Statistical analyses were ANOVA with Bonferroni's multiple comparison for BRET assays and ANOVA with Tukey’s post hoc test for densitometry and PLA assays;**p<0.01, ***p<0.001. Molecular weights indicated by arrows in blots.
-
Figure 3—figure supplement 5—source data 1
Figure 3—figure supplement 5 PLA assay of Beta-Arrestine1:ARHGAP21 interactions.
- https://doi.org/10.7554/eLife.24578.038
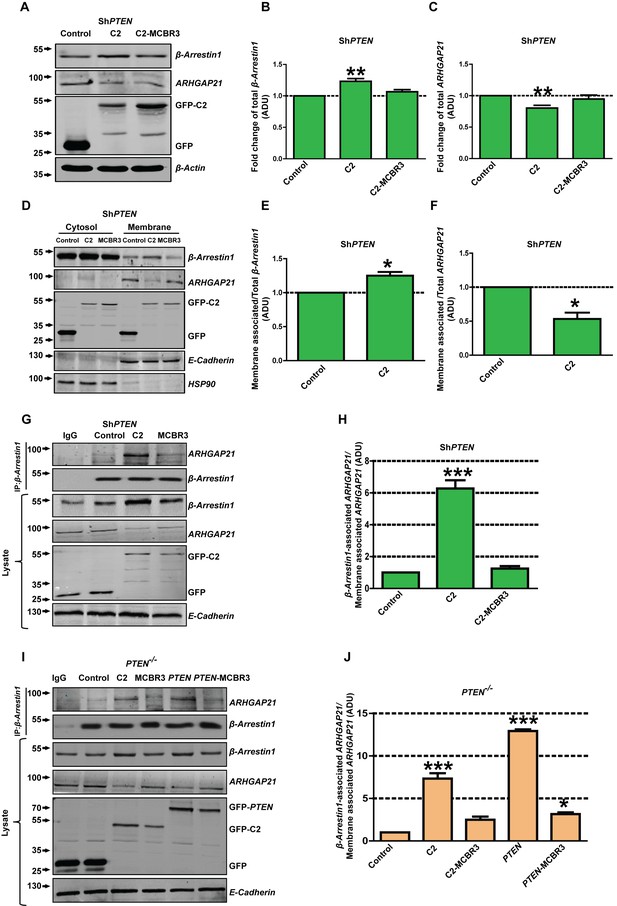
PTEN regulation of juxtamembrane β-Arrestin1 and ARHGAP21.
(A) shows total β-Arrestin1 and ARHGAP21 expression in Caco-2 ShPTEN cell lysates (top two panels) after PTEN C2 (C2), C2-MCBR3 or EV control transfections. Expression levels of transfected GFP-labelled proteins and β-Actin loading controls shown in lower two panels. (B) indicates summary fold changes of total β-Arrestin1; C2 = 1.23 ± 0.02; **p<0.01; MCBR3 = 1.07 ± 0.02; [MCBR3 vs control = NS]. (C) shows respective fold changes of ARHGAP21; C2 = 0.8 ± 0.02; **p<0.01; MCBR3 = 0.95 ± 0.03; [MCBR3 vs control = NS; all values in ADU]. (D) Effects of transfections on cytosol and membrane β-Arrestin1 and ARHGAP21 in Caco-2 ShPTEN (ShPTEN) cells (top two panels). Expression levels of transfected GFP-labelled proteins and E-Cadherin and HSP90 membrane and cytosolic markers shown in lower three panels. (E, F) Fold change of β-Arrestin1 and ARHGAP21 expression induced in the membrane fraction. (E) β-Arrestin1, C2 = 1.25 ± 0.05; *p=0.04 and (F) ARHGAP21 = 0.53 ± 0.09; *p=0.03. β-Arrestin1 and ARHGAP21 ADU values normalized against total lysate ADU for each protein. (G) β-Arrestin1-associated ARHGAP21 induced in ShPTEN membrane fractions by C2, C2-MCBR3 vs EV control transfections (top panel). β-Arrestin1 bait signal shown in second panel. Total β-Arrestin1 and ARHGAP21 in lysates shown in third and fourth panels. Expression of GFP-labeled C2, C2-MCBR3, EV and E-Cadherin membrane marker shown in lower three panels. (H) Fold changes of β-Arrestin1-associated ARHGAP21 normalized against total ARHGAP21 in the membrane fraction (ADU) - C2 = 6.27 ± 0.51; ***p<0.001; MCBR3 = 1.25 ± 0.15; [MCBR3 vs control = NS]. (I) β-Arrestin1-associated ARHGAP21 ADU after expression of C2, C2-MCBR3, wt PTEN or PTEN-MCBR3 vs EV control in PTEN -/- cell membrane fractions (top panel). β-Arrestin1 bait signal, total lysate expression of each protein, expression of GFP-labelled EV or C2 domain constructs and E-Cadherin membrane marker shown in lower five panels. (J) Summary fold changes of β-Arrestin1-associated ARHGAP21 normalized against total membrane ARHGAP21; C2 = 7.33 ± 0.64; C2-MCBR3 = 2.5 ± 0.35; PTEN = 12.93 ± 0.19; PTEN MCBR3 = 3.15 ± 0.21 ADU; control vs C2 or PTEN, ***p<0.001; control vs C2-MCBR3 (NS). Control vs PTEN-MCBR3 = *p<0.05. Analyses by ANOVA, Tukey post hoc or Student’s paired test. Molecular weights indicated by arrows in blots.
-
Figure 4—source data 1
Source data for Figure 4B.
- https://doi.org/10.7554/eLife.24578.046
-
Figure 4—source data 2
Source data for Figure 4C.
- https://doi.org/10.7554/eLife.24578.047
-
Figure 4—source data 3
Source data for Figure 4E.
- https://doi.org/10.7554/eLife.24578.048
-
Figure 4—source data 4
Source data for Figure 4F.
- https://doi.org/10.7554/eLife.24578.049
-
Figure 4—source data 5
Source data for Figure 4H.
- https://doi.org/10.7554/eLife.24578.050
-
Figure 4—source data 6
Source data for Figure 4J.
- https://doi.org/10.7554/eLife.24578.051
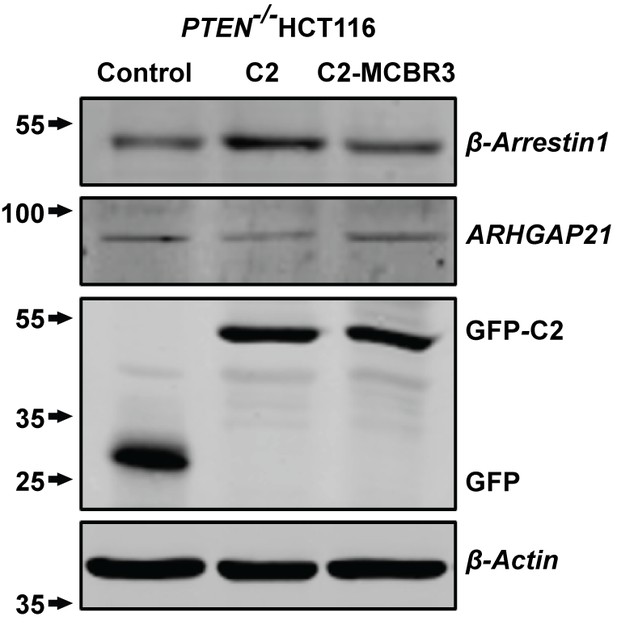
Total β-Arrestin1 and ARHGAP21 expression (top two panels) in.
PTEN -/- cell lysates after C2, C2-MCBR3 or EV control transfections. Expression levels of transfected GFP-labeled proteins and β-Actin loading controls shown in lower two panels.
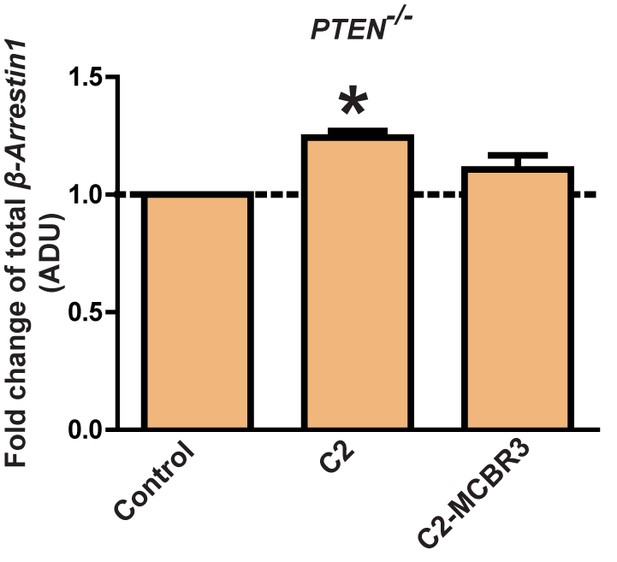
Fold changes of β-Arrestin1 - C2 = 1.24 ± 0.03; *p<0.05; C2-MCBR3 = 1.11 ± 0.06 [MCBR3 vs control = NS].
https://doi.org/10.7554/eLife.24578.041-
Figure 4—figure supplement 2—source data 1
Figure 4—figure supplement 2 Transfection effects on Beta-Arrestin1 in PTEN-/- cells.
- https://doi.org/10.7554/eLife.24578.052
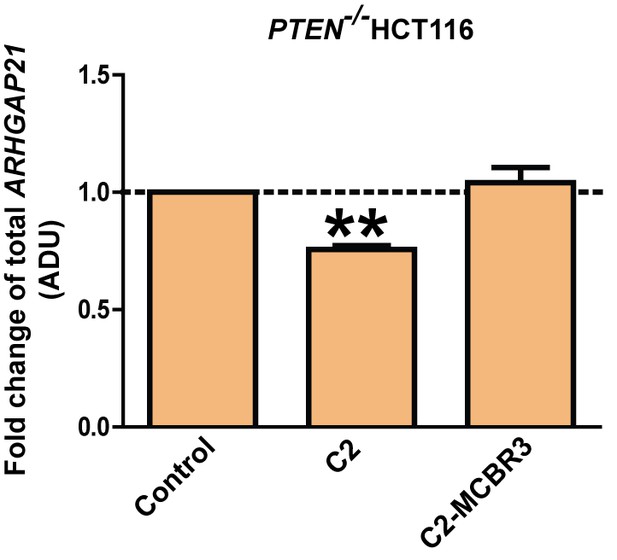
Fold changes of ARHGAP21.
C2 = 0.76 ± 0.02; **p<0.01; MCBR3 = 1.04 ± 0.06; [MCBR3 vs control = NS].
-
Figure 4—figure supplement 3—source data 1
Figure 4—figure supplement 3 Transfection effects on ARHGAP21 in PTEN-/- HCT116 cells.
- https://doi.org/10.7554/eLife.24578.053
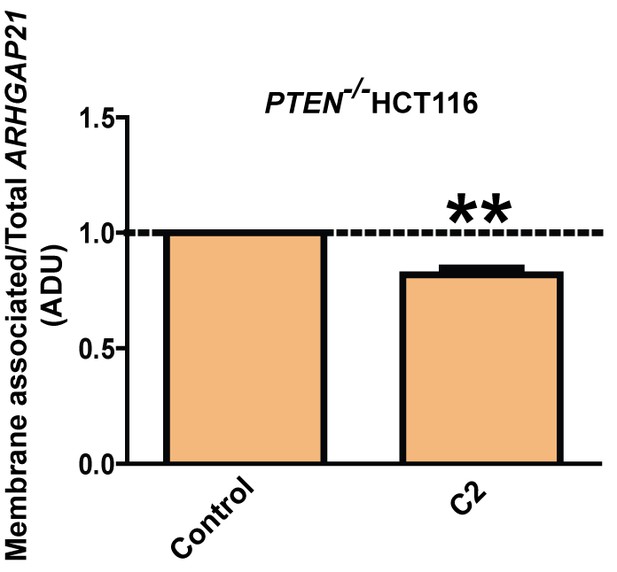
Fold changes of membrane ARHGAP21 (0.82 ± 0.03;**p<0.01).
ARHGAP21 expression values in membrane fractions were normalized against total expression of each protein. Molecular weights indicated by arrows in blots. Analyses by one-way ANOVA with Tukey post hoc test or Student’s paired t test.
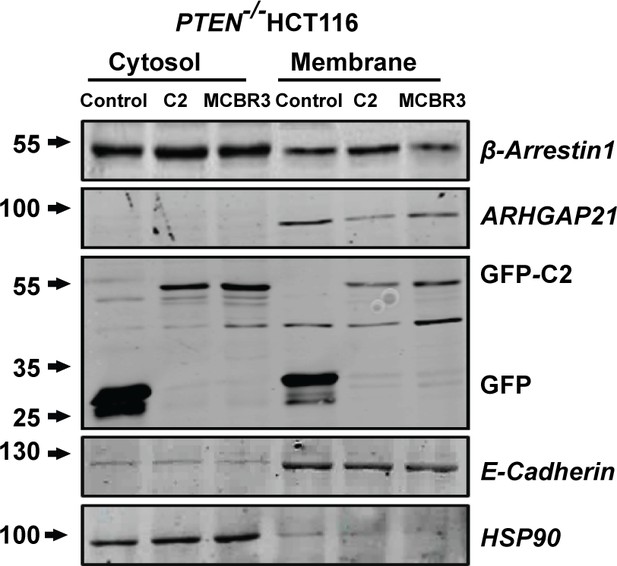
Effects of transfection on cytosol and membrane β-Arrestin1 and ARHGAP21 in PTEN -/- cells (top two panels).
Expression levels of transfected GFP-labeled proteins and E-Cadherin and HSP90 membrane and cytosolic markers shown in lower three panels.
-
Figure 4—figure supplement 5—source data 1
Figure 4—figure supplement 5 Membrane Beta-Arrestin1 in PTEN-/- cells - Source data.
- https://doi.org/10.7554/eLife.24578.054
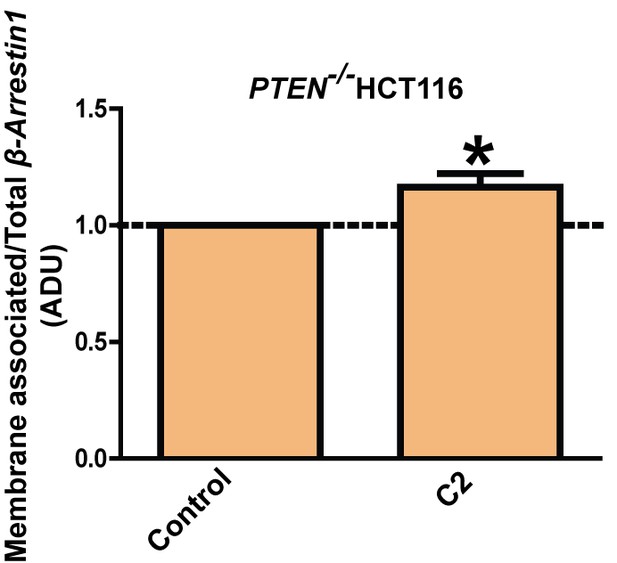
Fold changes of membrane β-Arrestin1 (1.16 ± 0.03; *p=0.04).
β-Arrestin1 expression values in membrane fractions were normalized against total expression of each protein. Molecular weights indicated by arrows in blots. Analyses by one-way ANOVA with Tukey post hoc test or Student’s paired t test.
-
Figure 4—figure supplement 6—source data 1
Figure 4—figure supplement 6 - Transfection effects on Membrane ARHGAP21in PTEN-/- cells.
- https://doi.org/10.7554/eLife.24578.055
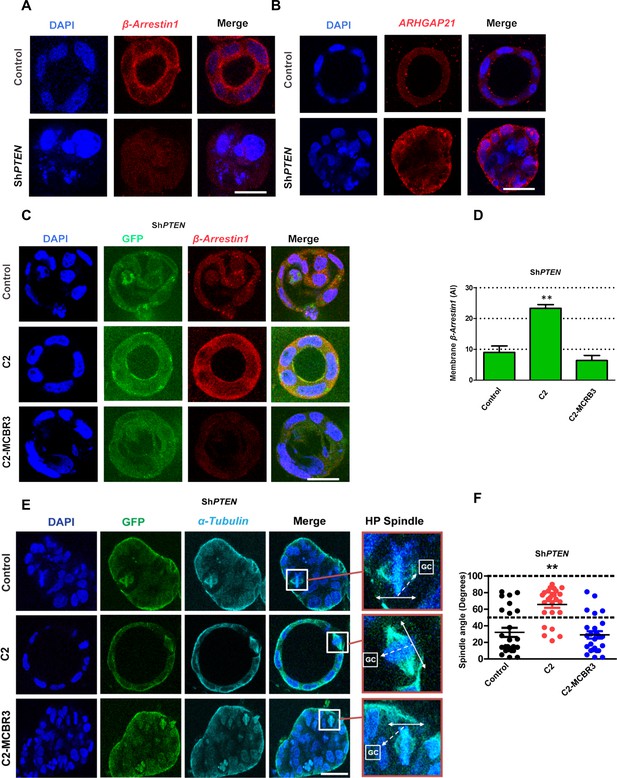
PTEN controls mitotic spindle orientation and 3D morphogenesis by noncatalytic regulation of β-Arrestin1.
(A) β-Arrestin1 and (B) ARHGAP21 immunofluorescence intensity in 3D control Caco-2 and ShPTEN cultures. (C) Effects of C2 vs C2-MCBR3 expression on membrane β-Arrestin1 immunoreactivity in ShPTEN cultures. (D) Summary cell membrane β-Arrestin1 immunoreactivity AI; EV control vs C2 vs C2-MCBR3 = 9.0 ± 2.08 vs 23.3 ± 1.20 vs 6.33 ± 1.67 AI;**p<0.01, control vs C2-MCBR3 = NS. (E) Effects of C2 vs C2-MCBR3 expression on spindle orientation. High-power (HP) spindle views (orange border) enlarge areas within white rectangles and show orientation angles (interrupted white arrows) of spindle planes (double-headed solid white arrows) toward gland centres (GCs). (F) Summary spindle angles relative to GCs in 3D ShPTEN cultures after transfection (● – Control = 32.08 ± 5.50 vs ● - C2 = 65.72 ± 4.10; **p<0.01 vs ● - MCBR3 = 29.1 ± 4.40; [MCBR3 vs control = NS], One-way ANOVA; **p<0.01, Tukey post hoc test. Imaging - DAPI [blue], β-Arrestin1 (A,C) red, ARHGAP21, (B) [red], GFP [green] and α-Tubulin (E) [cyan]. Scale bars = 20 µm.
-
Figure 5—source data 1
Source data for Figure 5D.
- https://doi.org/10.7554/eLife.24578.063
-
Figure 5—source data 2
Source data for Figure 5F.
- https://doi.org/10.7554/eLife.24578.064
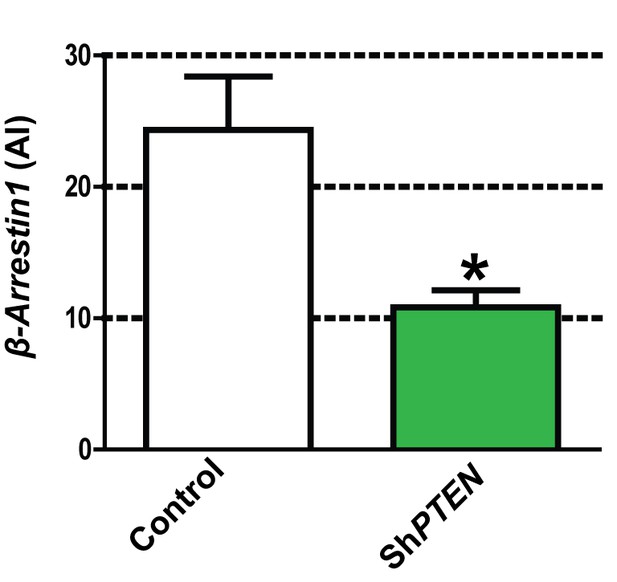
Summary β-Arrestin1 immunoreactivity (AI) in control Caco-2 vs ShPTEN organotypic cultures in Figure 5A = 24.3 ± 4.1 vs 11.00 ± 1.16;*p=0.04.
https://doi.org/10.7554/eLife.24578.057-
Figure 5—figure supplement 1—source data 1
Figure 5—figure supplement 1 - Beta-Arrestin 1 intensity in Caco-2 and ShPTEN.
- https://doi.org/10.7554/eLife.24578.065
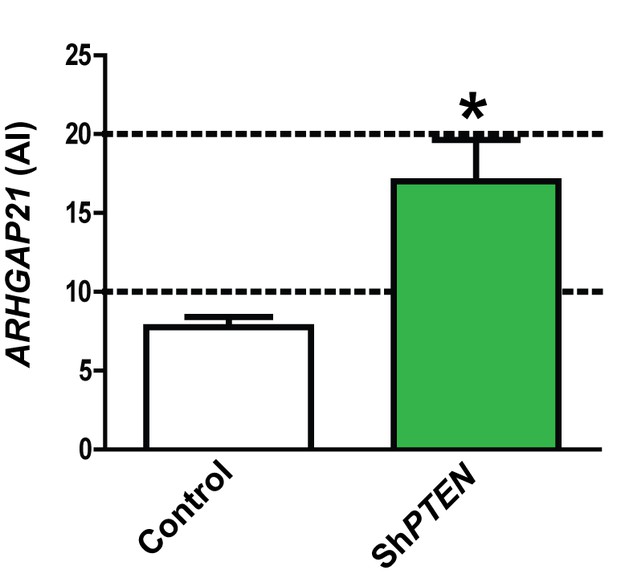
ARHGAP21 immunoreactivity (AI) in control Caco-2 vs ShPTEN organotypic cultures in Figure 5B = 8.0 ± 0.58 vs 17.0 ± 2.64 AI; *p=0.02.
https://doi.org/10.7554/eLife.24578.058-
Figure 5—figure supplement 2—source data 1
Figure 5—figure supplement 2 ARHGAP21 intensity in Caco-2 and ShPTEN glands.
- https://doi.org/10.7554/eLife.24578.066
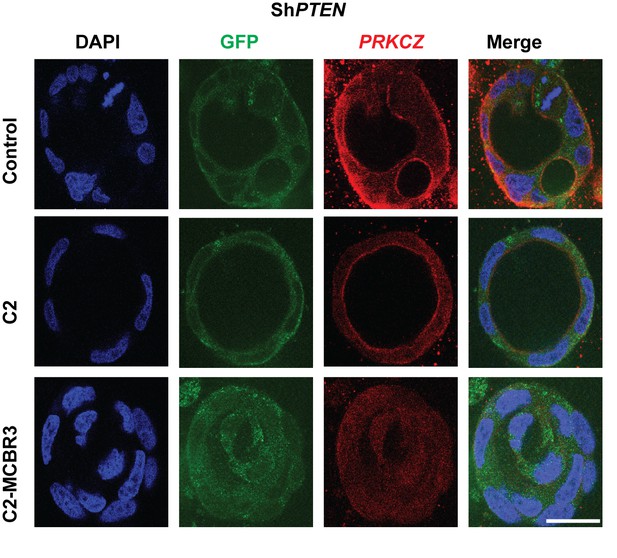
Effects of expression C2 or C2-MCBR3 vs EV control on lumen formation in ShPTEN cultures.
https://doi.org/10.7554/eLife.24578.059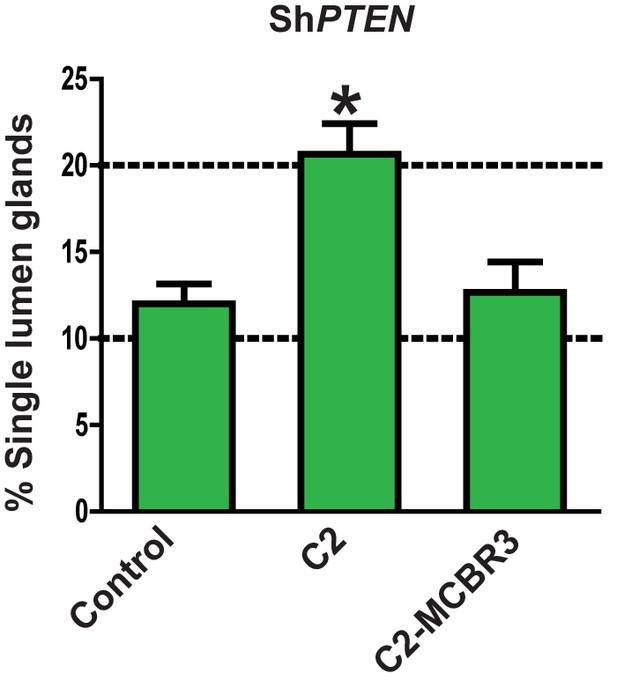
Effects of expression C2 or C2-MCBR3 vs EV control on lumen formation in ShPTEN cultures.
Indicates percentage ShPTEN glands with single central lumens after transfection, control = 12.0 ± 1.15%; C2 = 20.67 ± 1.76%; *p<0.05; MCBR3 = 12.67 ± 1.76%; [MCBR3 vs EV = NS].
-
Figure 5—figure supplement 4—source data 1
Figure 5—figure supplement 4 Transfection effects on single lumen formation in ShPTEN.
- https://doi.org/10.7554/eLife.24578.067
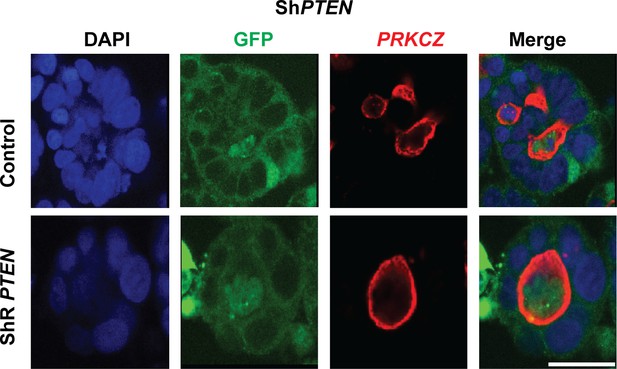
Effects of ShRNA-resistant (shR) PTEN or EV control on lumen formation in ShPTEN cultures.
https://doi.org/10.7554/eLife.24578.061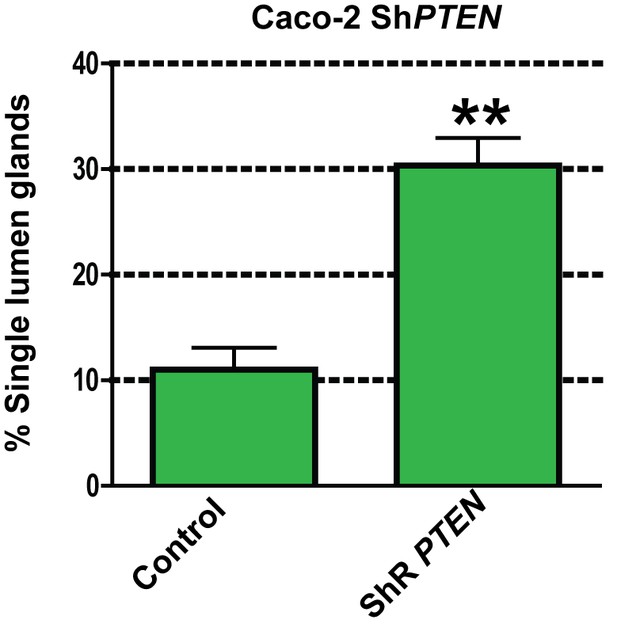
Summary effects of shR PTEN vs EV control on single lumen formation in 3D ShPTEN cultures - control = 11.33 ± 2.40%; ShR PTEN = 30.67 ± 2.91%; **p<0.01.
Analyses by ANOVA with Tukey post hoc test or Student’s t test. Imaging - DAPI (blue), GFP (green), protein kinase C zeta (PRKCZ), here used as an apical membrane marker (red). Scale bar 20 µm.
-
Figure 5—figure supplement 6—source data 1
Figure 5—figure supplement 6 Effects of shRNA resistant PTEN on single lumen formation in ShPTEN glands.
- https://doi.org/10.7554/eLife.24578.068
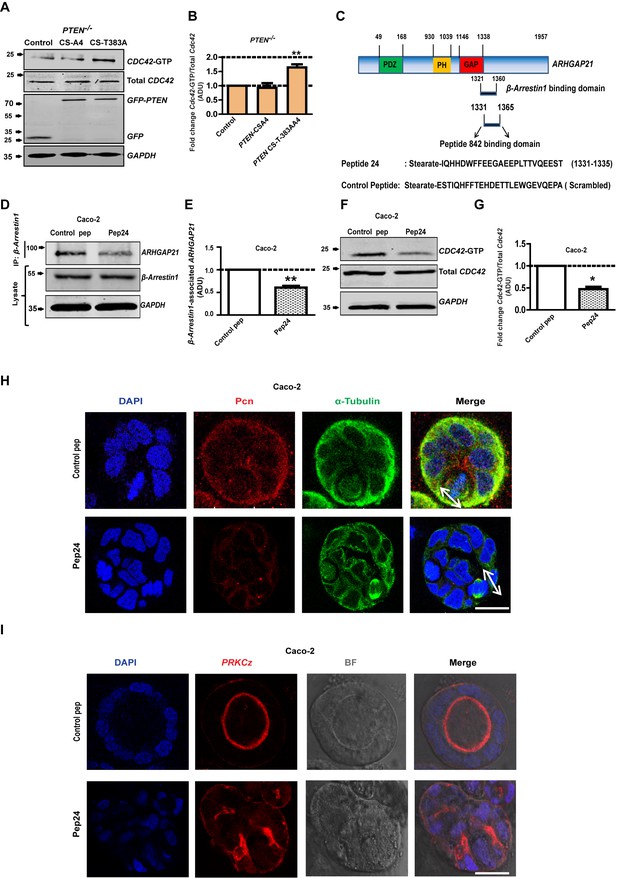
PTEN morphogenic functions mediated by β-Arrestin1, ARHGAP21 and Cdc42.
(A) Effects of PTEN CS-A4 or PTEN CS-T383A vs EV control on Cdc42 - GTP levels in PTEN -/- cells. (B) indicates fold change of Cdc42-GTP vs EV control; CS-A4 = 0.92 ± 0.16;p=NS; CS-T383A = 1.64 ± 0.11; **p=0.008. (C) Schematic of ARHGAP21 showing the β-Arrestin1-binding domain, the pep24 peptide-binding inhibitor and control peptide sequences (Anthony et al., 2011). Effects of pep24 vs control peptide (pep) on β-Arrestin1-ARHGAP21 binding (D,E) and Cdc42-GTP (F,G) in Caco-2 cells. (E) indicates fold change of β-Arrestin1-associated ARHGAP21 ADU in Caco-2 cells after pep24 vs control peptide treatment = 0.63 ± 0.02; **p=0.003. (G) indicates fold change of Cdc42-GTP in Caco-2 cells after pep24 vs control peptide treatment = 0.48 ± 0.05; **p=0.01. Treatment effects on spindle orientation (H) and lumenogenesis (I) in 3D Caco-2 cultures. Imaging for pericentrin (PCN) [red]; PRKCZ [red]; α-Tubulin [green], DAPI for nuclear DNA [blue] and bright-field (BF) imaging of lumen outlines. Spindle planes indicated by double-headed white arrows in Merge (H). Analyses by ANOVA, Tukey’s post hoc test. Scale bar - 20 µm. Molecular weights indicated by arrows in blots.
-
Figure 6—source data 1
. Figure 6B -Cdc42-GTP after transfection - source data
- https://doi.org/10.7554/eLife.24578.076
-
Figure 6—source data 2
Figure 6E - Peptide inhibitor treatment effects on Beta Arrestin1:ARHGAP21 binding in Caco-2
- https://doi.org/10.7554/eLife.24578.077
-
Figure 6—source data 3
Figure 6G Peptide inhibitor treatment effects on Cdc42-GTP in Caco-2
- https://doi.org/10.7554/eLife.24578.078
pep24 vs control peptide treatment effects on β-Arrestin1-associated ARHGAP21 ADU = 0.39 ± 0.09;*p=0.02.
https://doi.org/10.7554/eLife.24578.070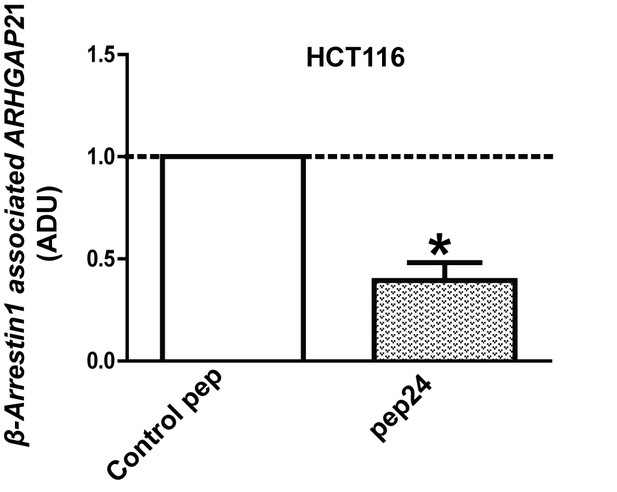
pep24 vs control peptide treatment effects on β-Arrestin1-associated ARHGAP21 ADU = 0.39 ± 0.09;*p=0.02.
https://doi.org/10.7554/eLife.24578.071-
Figure 6—figure supplement 2—source data 1
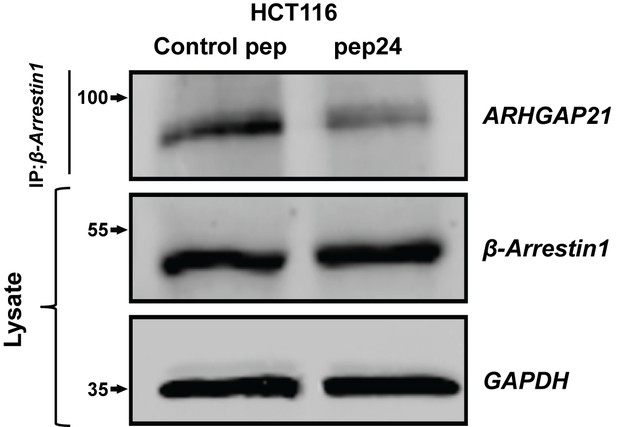
Figure 6—figure supplement 2 Effects of peptide binding inhibitor on Beta-arrestin1:ARHGAP21 interactions in HCT116 cells
- https://doi.org/10.7554/eLife.24578.079
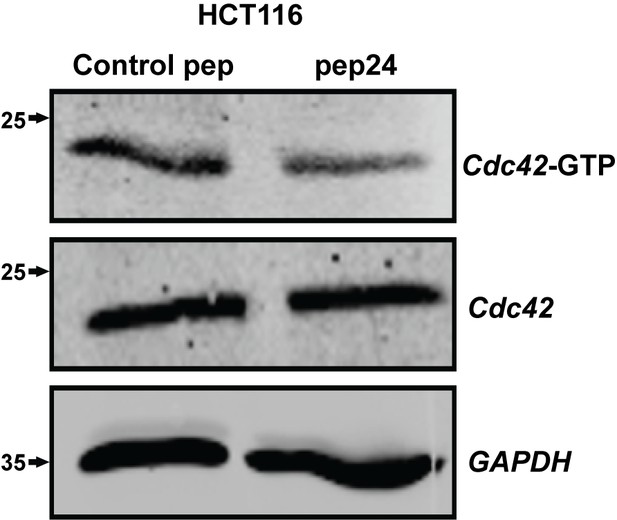
pep24 vs control peptide treatment effects on Cdc42-GTP ADU = 0.44 ± 0.02; **p<0.01 in HCT116 cells.
https://doi.org/10.7554/eLife.24578.072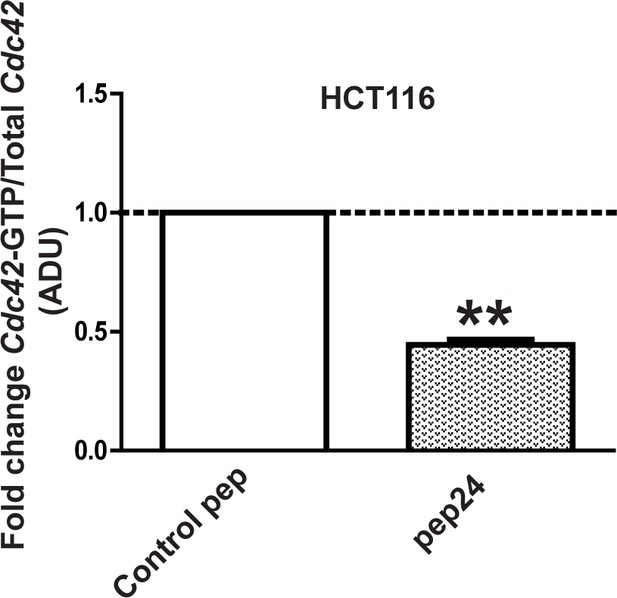
pep24 vs control peptide treatment effects on Cdc42-GTP ADU = 0.44 ± 0.02; **p<0.01 in HCT116 cells.
https://doi.org/10.7554/eLife.24578.073-
Figure 6—figure supplement 4—source data 1
Figure 6—figure supplement 4 - Peptide inhibitor treatment effects in Cdc42-GTP in HCT116 cells - source data
- https://doi.org/10.7554/eLife.24578.080
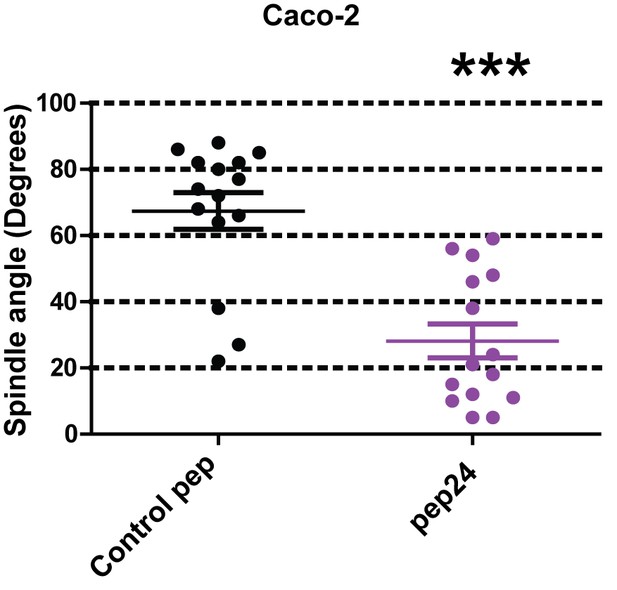
Summary treatment effects on mitotic spindle angles [●Control peptide = 67.4 ± 5.50 vs ● pep24 = 28.1 ± 5.10; ***p<0.001] and % glands with single central lumens in 3D Caco-2 cultures (Control peptide = 35.33 ± 1.76% vs pep24 = 20.67 ± 1.76%;**p<0.01).
Molecular weights indicated by arrows in blots. Analysis by paired Student’s t test.
-
Figure 6—figure supplement 5—source data 1
Figure 6—figure supplement 5 - Peptide inhibitor treatment effects on spindle angles in Caco-2 cultures
- https://doi.org/10.7554/eLife.24578.081
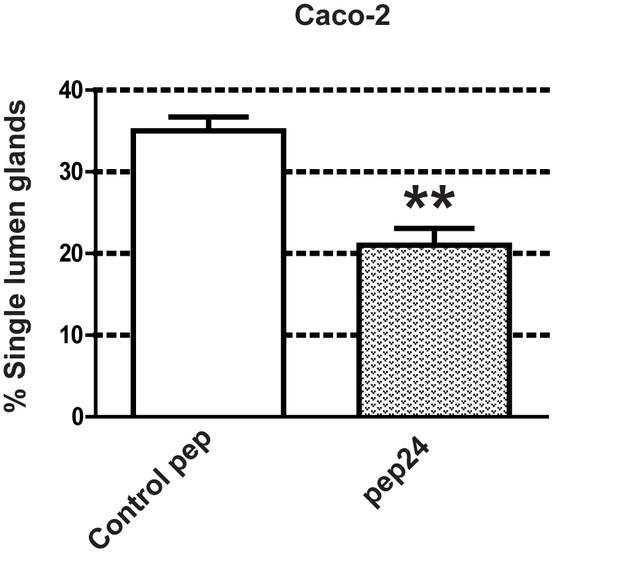
Summary treatment effects on mitotic spindle angles [●Control peptide = 67.4 ± 5.50 vs ● pep24 = 28.1 ± 5.10; ***p<0.001] and % glands with single central lumens in 3D Caco-2 cultures (Control peptide = 35.33 ± 1.76% vs pep24 = 20.67 ± 1.76%;**p<0.01).
Molecular weights indicated by arrows in blots. Analysis by paired Student’s t test.
-
Figure 6—figure supplement 6—source data 1
Figure 6—figure supplement 6 Peptide inhibitor treatment effects on spindle angles in Caco-2 cultures
- https://doi.org/10.7554/eLife.24578.082
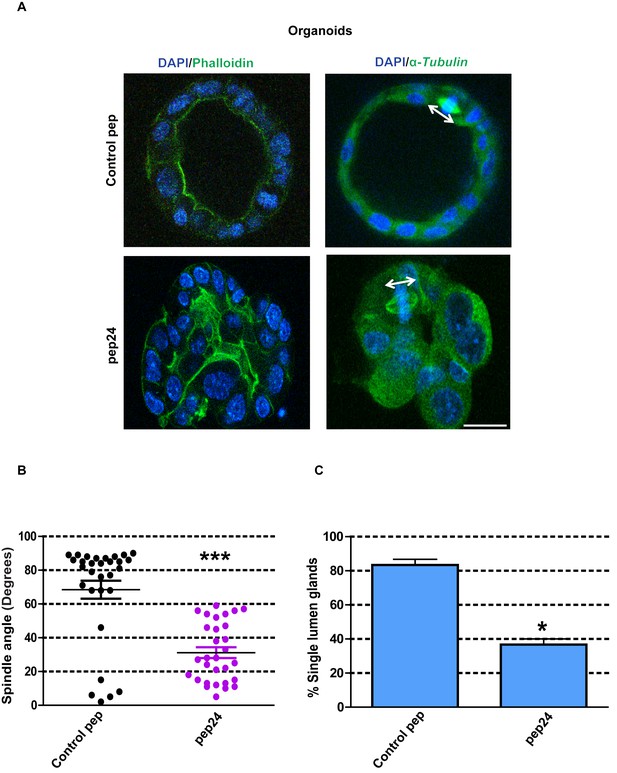
β-Arrestin1-ARHGAP21 binding is essential for morphogenesis of normal colorectal organoids.
(A) Effects of pep24 vs control peptide (pep) on multicellular morphogenesis of normal colorectal epithelium. Imaging - apical actin marker FITC-labeled phalloidin [green], DAPI for nuclear DNA [blue] and FITC-labeled α-Tubulin [green]. Spindle orientation indicated by double-headed white arrows. (B) Summary spindle angles in 3D colorectal organoids after treatment (n = 30 per treatment group) ●Control peptide = 68.47 ± 5.370 vs ●pep24 = 31.13 ± 3.200; ***p<0.001. (C) Summary data represent percentage glandular colorectal organoids with single central lumens after treatment; control peptide = 80.00 ± 5.77% vs pep24 = 36.67 ± 3.3%;*p=0.039; (n = 10 organoids per treatment group in triplicate). Student’s t test. Scale bar 20 µm.
-
Figure 7—source data 1
Figure 7B Spindle angles in organoids - souce data.
- https://doi.org/10.7554/eLife.24578.084
-
Figure 7—source data 2
Figure 7C Single central lumen formation in organoids - source data.
- https://doi.org/10.7554/eLife.24578.085
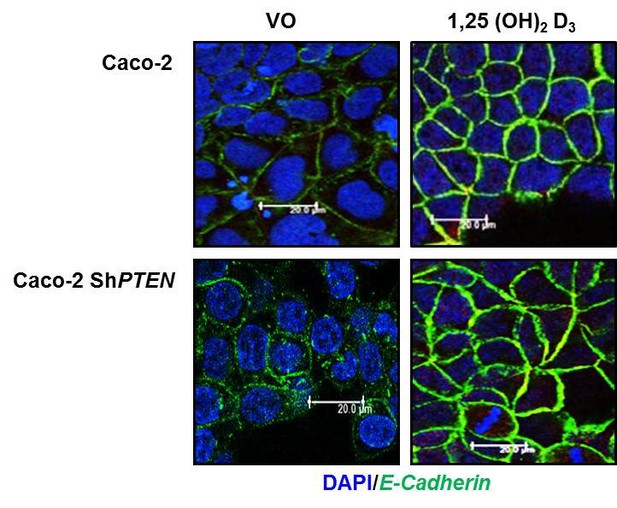
To investigate effects of PTEN on ligand- mediated enhancement of a membrane protein (Reviewer 2, point 2, Figure 1C), we tested 100nM 1,25 (OH)2 Vit D3vs vehicle only control against membrane recruitment of E-Cadherin (4) in Caco-2 and Caco-2 ShPTEN cells.